Identification of aluminium transport-related genes via genome-wide phenotypic screening of Saccharomyces cerevisiae†
Abstract
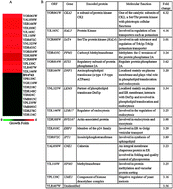
Genome-wide screening using gene deletion mutants has been widely carried out with numerous toxicants including oxidants and metal ions. The focus of such studies usually centres on identifying sensitive phenotypes against a given toxicant. Here, we screened the complete collection of yeast gene deletion mutants (5047) with increasing concentrations of aluminium sulphate (0.4, 0.8, 1.6 and 3.2 mM) in order to discover aluminium (Al3+) tolerant phenotypes. Fifteen genes were found to be associated with Al3+ transport because their deletion mutants exhibited Al3+ tolerance, including lem3Δ, hal5Δ and cka2Δ. Deletion of CKA2, a catalytic subunit of tetrameric protein kinase CK2, gives rise to the most pronounced resistance to Al3+ by showing significantly higher growth compared to the wild type. Functional analysis revealed that both molecular regulation and endocytosis are involved in Al3+ transport for yeast. Further investigations were extended to all the four subunits of CK2 (CKA1, CKA2, CKB1 and CKB2) and the other 14 identified mutants under a spectrum of metal ions, including Al3+, Zn2+, Mn2+, Fe2+, Fe3+, Co3+, Ga3+, Cd2+, In3+, Ni2+ and Cu2+, as well as hydrogen peroxide and diamide, in order to unravel cross-tolerance amongst metal ions and the effect of the oxidants. Finally, the implication of the findings in Al3+ transport for the other species like plants and humans is discussed.

 Please wait while we load your content...
Please wait while we load your content...