Insights into the structural basis of 3,5-diaminoindazoles as CDK2 inhibitors: prediction of binding modes and potency by QM–MM interaction, MESP and MD simulation†
Abstract
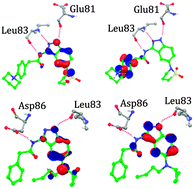
The novel 3,5-diaminoindazole derivatives are well-known as potent and anti-proliferative cyclin-dependent kinase 2 inhibitors. We report a combined quantum mechanics/molecular mechanics study to determine the protein–ligand interaction energy, and some quantum chemical descriptors to successfully rank these inhibitors. The results in this work show that the QM–MM interaction energy is strongly correlated to the biological activity and can be used as a predictor, which was further validated by Spearman's rank correlation coefficient. An exhaustive analysis of the protein–ligand structures obtained from molecular dynamics simulations shows specific interactions within the active site. Furthermore, the docking study was supported by electronic property analysis using density functional theory at the B3LYP/3-21*G level. The results obtained from molecular docking and surface analysis shed some insight on steric and electronic complementarities of these molecules to CDK2. Aqueous solvation energy values give an indication of the solubility and can be used as a guide for the pharmacokinetic optimization of these molecules. Furthermore, ADME/T properties calculated are in the desirable range, so these compounds are predicted to be drug like with low toxicity potential. Overall, the approach was successful in the cases considered, and it could be useful for the design of inhibitors in the lead optimization phase of drug discovery against CDK2.

 Please wait while we load your content...
Please wait while we load your content...