Understanding the drug resistance mechanism of hepatitis C virus NS5B to PF-00868554 due to mutations of the 423 site: a computational study
Abstract
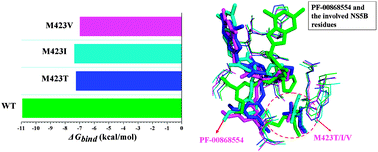
NS5B, a hepatitis C virus (HCV) RNA-dependent RNA polymerase (RdRp) that plays a key role in viral replication, is an important target in the discovery of antiviral agents. PF-00868554 is a potent non-nucleoside inhibitor (NNI) that binds to the Thumb II allosteric pocket of NS5B polymerase and has shown significant promise in phase II clinical trials. Unfortunately, several PF-00868554 resistant mutants have been identified. M423 variants were the most common NS5B mutations that occurred after PF-00868554 monotherapy. In this study, we used molecular dynamics (MD) simulations, binding free energy calculations and free energy decomposition to explore the drug resistance mechanism of HCV to PF-00868554 resulting from three representative mutations (M423T/V/I) in NS5B polymerase. Free energy decomposition analysis reveals that the loss of binding affinity mainly comes from the reduction of both van der Waals (ΔEvdw) and electrostatic interaction contributions in the gas phase (ΔEele). Further structural analysis indicates that the location of PF-00868554 and the binding mode changed due to mutation of the residue at the 423 site of NS5B polymerase from methionine to threonine, isoleucine or valine, which further resulted in the loss of binding ability of PF-00868554 to NS5B polymerase. The obtained computational results will have important value for the rational design of novel non-nucleoside inhibitors targeting HCV NS5B polymerase.

 Please wait while we load your content...
Please wait while we load your content...