Label-free fluorescence detection of DNA methylation and methyltransferase activity based on restriction endonuclease HpaII and exonuclease III
Abstract
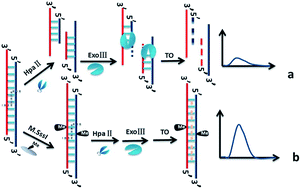
Strategies to detect the methylation of site specific DNA and assay of M.SssI methyltransferase (M.SssI MTase) activity are important in determining human cancers due to aberrant methylation linked to cancer initiation and progression. Herein, we report a label-free fluorescence detection method for DNA methylation and MTase activity based on restriction endonuclease HpaII and exonuclease III (Exo III). A label-free probe DNA was designed, which hybridized with target DNA (one 32-mer DNA from the exon 8 promoter region of the Homo sapiens p53 gene) to form double stranded DNA (dsDNA). Upon the cleavage action of HpaII and degradation reaction of Exo III, dsDNA changed to single stranded DNA (ssDNA) and the fluorescence intensity of thiazole orange (TO) is weak. After the resulting dsDNA was methylated by M.SssI MTase, the action of HpaII and Exo III was prevented, then TO intercalates into the dsDNA and emits strong fluorescence. This method can determine DNA methylation at the site of CpG and distinguish a one-base mismatched target sequence. The fluorescence intensity has a linear relationship with M.SssI MTase activities in the range of 1–10 U mL−1 with a detection limit of 0.16 U mL−1 in terms of 3 times deviation of the blank sample. The methylation of DNA by a hydroxyl radical triggered by DMSO and CH3CHO was also measured. These results show that the proposed method can specifically and selectively detect DNA methylation and M.SssI MTase activity. Human serum has no obvious effects on the assay performance, indicating that the method has great potential for further application in complex samples.

 Please wait while we load your content...
Please wait while we load your content...