Label-free electrochemiluminescence detection of specific-sequence DNA based on DNA probes capped ion nanochannels†
Abstract
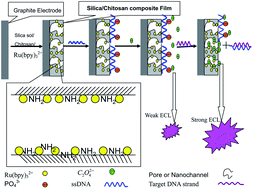
As one of the powerful molecular recognition elements, the functional DNA probes have been successfully utilized to construct various biosensors. However, the accurate readout of the recognition event of DNA probe binding to the specific target by label-free means is still challenging. Here, a simple and label-free electrochemiluminescence (ECL) method for sensing the recognition event of DNA probe to sequence-specific DNA is developed. Oxalate is used as an ECL co-reactant and p53 tumor suppressor gene as a model of target analyte. In the ECL sensing platform, the nanochannel structural film, which contains silica-sol, chitosan and Ru(bpy)32+, is prepared by an electrochemical deposition method. Then, DNA probes are attached onto the surface of the nanochannel-based composite film electrode based on the stronger interaction between DNA probes and chitosan embedded in the ECL composite film. These nanochannels were capped by the DNA probes. As a result, the mass-transfer channel between the Ru(bpy)32+ embedded in the nanochannel-based composite film and the ECL co-reactant in the bulk solution was greatly blocked and a weak ECL signal was observed. Conversely, in the presence of target sequences, the hybridizing reaction of targets with DNA probes could result in the escape of the DNA probes from the composite film due to the rigid structure of the duplex DNA. Thus, these nanochannels were uncapped and a stronger ECL signal was detected. Our results show that this ECL method could effectively discriminate complementary from single-base mismatch DNA sequences. Under the optimal conditions, the linear range for target DNA was from 1.0 × 10−11 to 1.0 × 10−9 mol L−1 with a detect limit of 2.7 × 10−12 mol L−1. This work demonstrates that porous structures on the silica–chitosan composite film can provide a label-free and general platform to measure the change of DNA configuration.

 Please wait while we load your content...
Please wait while we load your content...