Putative cobalt- and nickel-binding proteins and motifs in Streptococcus pneumoniae†
Abstract
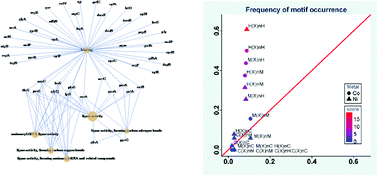
Cobalt and nickel play important roles in various biological processes. The present work focuses on the enrichment and identification of Co- and Ni-binding motifs and proteins in Gram-positive bacteria. Immobilized metal affinity column (IMAC) was used to partially enrich putative metal-binding proteins and peptides from Streptococcus pneumoniae, and then LTQ-Orbitrap mass spectrometry (MS) was applied to identify and characterize the metal-binding motifs and proteins. In total, 208 and 223 proteins were isolated by Co- and Ni-IMAC columns respectively, in which 129 proteins were present in both preparations. Based on the gene ontology (GO) analysis, the putative metal-binding proteins were found to be mainly involved in protein metabolism, gene expression regulation and carbohydrate metabolism. These putative metal-binding proteins form a highly connected network, indicating that they may synergistically work together to achieve specific biological functions. Putative Co- and Ni-binding motifs were identified with H(X)nH, M(X)nH and H(X)nM derived from the identified 51 Co-binding peptides and 66 Ni-binding peptides. Statistics of frequency of amino acids in the metal-binding motifs showed that cobalt and nickel prefer to bind histidine and methionine, but not cysteine. These results obtained by a systematic metalloproteomic approach provide important clues for the further investigation of metal homeostasis and metal-related virulence of bacteria.
- This article is part of the themed collections: Nickel in biology and Metallomics in China

 Please wait while we load your content...
Please wait while we load your content...