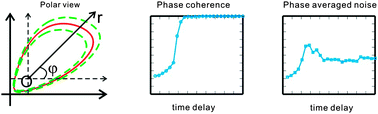
The molecular segmentation clock is a complex regulatory network that governs the periodic somite segmentation in vertebrate embryos. Underlying the rhythm of the segmentation clock is a single-cell level pace-making circuit, where inevitable molecular noise and time delay impose normal operating constraints to the pace-making. However, how the molecular mechanisms of the core circuit of the segmentation clock coordinate the operating constraints and maintain the rhythmic nature of the developmental process remains poorly understood. To address this question, we construct two biologically-motivated mathematical models with multiple clock protein transcription binding sites, with differential or rate-limited decay rates for protein monomers and dimers. We demonstrate that the rate-limited decay significantly enlarges the parameter space of noise-induced and delay-induced oscillations. Interestingly, focusing on the stochastic characters of noise-induced and delay-induced oscillations in terms of phase coherence and phase averaged amplitude noise in the polar coordinate, we find that there is a delay-managed tradeoff between phase coherence and phase averaged amplitude noise. In particular, the model with both multiple binding sites and rate-limited decay can show regular tunability as the delay increases. Our results indicate that transcriptional and post-translational mechanisms constrain the combined effects of noise and delay on the molecular dynamics of the segmentation clock.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...