Network function shapes network structure: the case of the Arabidopsis flower organ specification genetic network†
Abstract
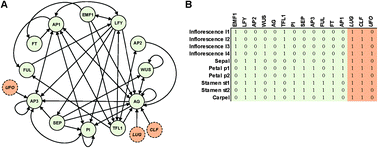
The reconstruction of many biological networks has allowed detailed studies of their structural properties. Several features exhibited by these networks have been interpreted to be the result of evolutionary dynamics. For instance the degree distributions may follow from a preferential attachment of new genes to older ones during evolution. Here we argue that even in the absence of any evolutionary dynamics, the presence of atypical features may follow from the fact that the network implements certain functions. To examine this network function shapes network structure scenario, we focus on the Arabidopsis genetic network controlling early flower organogenesis in which gene expression dynamics has been modelled using a Boolean framework. Specifically, for a system with 15 master genes, the phenotype consists of 10 experimentally determined steady-state expression patterns, considered here as the functional constraints on the network. The space of genetic networks satisfying these constraints is sometimes referred to as the neutral or genotype network. We sample this space using Markov Chain Monte Carlo which allows us to demonstrate how the functional (phenotypic) constraints shape the gene network structure. We find that this shaping is strongest for the edge (interaction) usage, with effects that are functionally interpretable. In contrast, higher order features such as degree assortativity and network motifs are hardly shaped by the phenotypic constraints.
- This article is part of the themed collection: Molecular BioSystems Network Biology

 Please wait while we load your content...
Please wait while we load your content...