Reconstruction and analysis of the genome-scale metabolic network of Candida glabrata†
Abstract
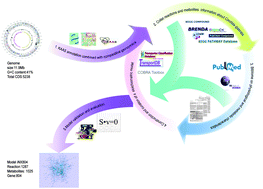
Candida glabrata has been recognized as an efficient industrial microorganism for the production of pyruvate, the most widely used α-oxocarboxylic acid in the areas of agrochemicals, drugs, and chemicals. To gain a comprehensive understanding of its physiology and cellular metabolism, the genome-scale metabolic model for C. glabrata (named iNX804), comprising 804 genes, 1287 reactions, and 1025 metabolites, was reconstructed by genome sequence annotation and biochemical data mining. The flux balance analysis data of model iNX804 exhibited good agreement with the experimental data, including the phenotypic data and the carbon flux distribution. The model iNX804 predicted that C. glabrata can synthesize a high concentration of pyruvate because it has a strong glucose transport capacity and three pyruvate biosynthesis pathways and is deficient in key steps in vitamin synthesis. Furthermore, the metabolic capacity of pyruvate and other fine chemicals derived from pyruvate was evaluated by in silico deletion and overexpression of several key genes. The metabolic model iNX804 provides a potential platform for global elucidation of the metabolism of C. glabrata.

 Please wait while we load your content...
Please wait while we load your content...