Rapid high-throughput genotyping of HBV DNA using a modified hybridization-extension technique
Abstract
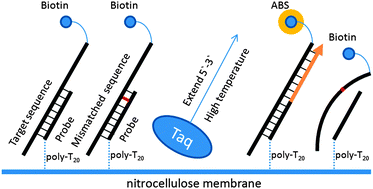
China has the highest incidence of hepatitis B virus (HBV) infection worldwide. HBV genotypes have variable impacts on disease pathogenesis and drug tolerance. We have developed a technically simple and accurate method for HBV genotyping that will be applicable to pre-treatment diagnosis and individualized treatment. Multiple sequence alignments of HBV genomes from GenBank were used to design primers and probes for genotyping of HBV A through H. The hybridization was carried out on nitrocellulose (NC) membranes with probes fixed in an array format, which was followed by hybrid amplification by an extension step with DNA polymerase to reinforce the double-stranded DNA hybrids on the NC membrane and subsequent visualization using an avidin–biotin system. Genotyping results were confirmed by DNA sequencing and bioinformatics analysis using the National Center for Biotechnology Information genotyping database, and compared with results from the line probe assay. The data show that multiple sequence alignment defined a 630 bp region in the HBV PreS and S regions that was suitable for genotyping. All genotyping significant single nucleotides in the region were defined. Two-hundred-and-ninety-one HBV-positive serum samples from Northwest Chinese patients were genotyped, and the genotyping rate from the new modified hybridization-extension method was 100% compared with direct sequencing. Compared with line probe assay, the newly developed method is superior, featuring reduced reaction time, lower risk of contamination, and increased accuracy for detecting single nucleotide mutation. In conclusion, a novel hybridization-extension method for HBV genotyping was established, which represents a new tool for accurate and rapid SNP detection that will benefit clinical testing.
- This article is part of the themed collection: Coronavirus articles - free to access collection

 Please wait while we load your content...
Please wait while we load your content...