Computational screening of one- and two-photon spectrally tuned channelrhodopsin mutants†
Abstract
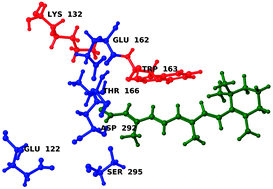
Optogenetics is by now a well-established field within neuroscience where neuro-response is controlled at the molecular level using the photochemical properties of channelrhodopsin (ChR). In this study the recently published X-ray structure of retinal inside the ChR binding pocket serves as the basis for conducting high-level polarizable embedding quantum mechanical/molecular mechanical (QM/MM) mutation studies with the aim of providing insight into the tuning mechanisms of this remarkable protein. The levels of theory applied are the recently developed PERI-CC2 and PE-DFT approaches. Their computational efficiency makes it possible to rapidly carry out a large number of spectral calculations. This is exploited to construct in silico mutated ChR variants which are characterized in terms of the location of the relevant excitation energy and the magnitude of the two-photon absorption cross section. In turn, this allows us to pinpoint the amino acids that have the largest electrostatic effect on the studied excited state properties. We show that a single/double site mutation strategy in ChR does not perturb the electronic properties of retinal to a degree that satisfies the experimental demand for a significant red-shift. With respect to non-linear absorption we conjecture that the recently synthesized ChETA variant possesses an even larger two-photon cross section than the C1C2 variant and it is thus an ideal candidate for further studies on the two-photon activation of ChR.

 Please wait while we load your content...
Please wait while we load your content...