QM/MM simulations of vibrational spectra of bacteriorhodopsin and channelrhodopsin-2†
Abstract
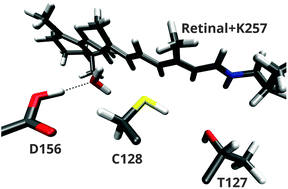
Channelrhodopsin-2 is a light-gated ion channel, which has been studied intensively over the last decade. Vibrational spectroscopic experiments started to shed light on the structural changes, that occur during the photocycle, especially in the hydrogen-bonded network surrounding the protonated D156 and C128 – the DC gate. However, the interpretation of these experiments was only based on homology models. Since then, an X-ray structure and better computational models became available. In this article, we show that in combination with a recent reparametrization, the approximate DFT method, DFTB, is able to describe the effects of hydrogen bonding on the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch vibration in
O stretch vibration in
- This article is part of the themed collection: Bunsentagung 2013: 'Theory meets Spectroscopy'

 Please wait while we load your content...
Please wait while we load your content...