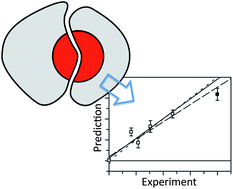
We have successfully developed and validated with experiment a new computational method for quantitatively predicting the effects of mutations at a protein–protein interface. From over 500 ns of explicitly solvated molecular dynamics, Mutational Locally Enhanced Sampling (MULES) shows significantly improved accuracy over post-processing methods for a prototypical set of mutations, with a maximum mean unsigned error to experiment of 0.5 kcal mol−1 and comparable or better precision. The technique in principle allows the effect of any mutation to be calculated, whether natural or non-natural. The versatility, quantitative accuracy, high precision and speed of MULES compared to existing computational prediction techniques enhance its potential for modelling changes to the interface in a systematic way, thereby aiding peptide and protein interaction design.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?

 Please wait while we load your content...
Please wait while we load your content...