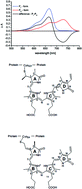
The plant pathogen Pseudomonas syringae pv. tomato carries two genes encoding bacterial phytochromes. Sequence motifs identify both proteins (PstBphP1 and PstBphP2, respectively) as biliverdin IXα (BV)-binding phytochromes. PstbphP1 is arranged in an operon with a heme oxygenase (PstBphO)-encoding gene (PstbphO), whereas PstbphP2 is flanked downstream by a gene encoding a CheY-type response regulator. Expression of the heme oxygenase PstBphO yielded a green protein (λmax = 650 nm), indicative for bound BV. Heterologous expression of PstbphP1 and PstbphP2 and in vitro assembly with BV IXα yielded the apoproteins for both phytochromes, but only in the case of PstBphP1 a light-inducible chromoprotein. Attempts to express the endogenous heme oxygenase BphO and either of the two phytochromes from two plasmids yielded only holo-PstBphP1. Relatively small amounts of soluble holo-PstBphP2 were just obtained upon co-expression with BphO from P. aeruginosa. Expression of the operon containing PstbphO:PstbphP1 led to an improved yield and better photoreactivity for PstBphP1, whereas an identical construct, exchanging PstbphP1 for PstbphP2 (PstbphO:PstbphP2), again yielded only minute amounts of chromophore-loaded BphP2-holoprotein. The improved yield for PstBphP1 from the PstbphO:PstbphP1 operon expression is apparently caused by complex formation between both proteins during biosynthesis as affinity chromatography of either protein using two different tags always co-purified the reaction partner. These results support the importance of protein–protein interactions during tetrapyrrole metabolism and phytochrome assembly.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...