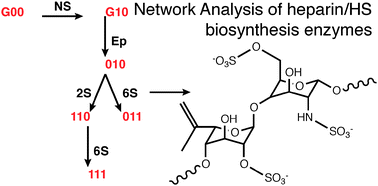
The form of the biosynthetic pathway of the biologically and medically important polysaccharides heparan sulfate (HS) and the closely related heparin remain obscure despite significant progress characterising the biosynthetic machinery. Considering possible biosynthetic schemes using a graph approach and applying known constraints of enzyme order and specificity, a previously unreported system with a highly efficient tree structure emerged with two features: (1) All commonly occurring HS disaccharides could be synthesised through a common route, the major branch. (2) The least common disaccharides also occurred on a separate common branch, termed here the minor branch. This suggested that the relative abundance of these two sets of structures were the result of the specificity of a single enzyme (HS epimerase) acting at an early point in the scheme, to convert GlcA-GlcNS to IdoA-GlcNS in preference to converting GlcA-GlcNAc to IdoA-GlcNAc. A third key finding was that the common substrates for 3-O-sulfation all lie on the same (major) branch. The proposed scheme is consistent with a wide body of experiments comprising both biochemical data and results from HS biosynthetic enzyme knockout experiments in the literature. The major branch also contains a bifurcation, providing a choice of two distinct backbone geometries with the same charge. Further development of this novel biosynthetic scheme, in which frame shifts in the site of action of the enzymes were permitted to occur, while maintaining their order of action, suggested a mechanism by which domains could be generated, or further modification blocked. The relationship between the proposed pathway and the geometric and charge possibilities it allows were also explored.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...