Optimality and thermodynamics determine the evolution of transcriptional regulatory networks†
Abstract
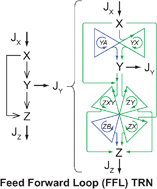
Transcriptional motifs are small regulatory interaction patterns that regulate biological functions in highly-interacting cellular networks. Recently, attempts have been made to explain the significance of transcriptional motifs through dynamic function. However, fundamental questions remain unanswered. Why are certain transcriptional motifs with similar dynamic function abundant while others occur rarely? What are the criteria for topological generalization of these motifs into complex networks? Here, we present a novel paradigm that combines non-equilibrium thermodynamics with multiobjective-optimality for network analysis. We found that energetic cost, defined herein as specific dissipation energy, is minimal at the optimal environmental conditions and it correlates inversely with the abundance of the network motifs obtained experimentally for E. coli and S. cerevisiae. This yields evidence that dissipative energetics is the underlying criteria used during evolution for motif selection and that biological systems during transcription tend towards evolutionary selection of subgraphs which produces minimum specific heat dissipation under optimal conditions, thereby explaining the abundance/rare occurrence of some motifs. We show that although certain motifs had similar dynamical functionality, they had significantly different energetic cost, thus explaining the abundance/rare occurrence of these motifs. The presented insights may establish global thermodynamic analysis as a backbone in designing and understanding complex networks systems, such as metabolic and

 Please wait while we load your content...
Please wait while we load your content...