Statistics-based model for basis set superposition error correction in large biomolecules
Abstract
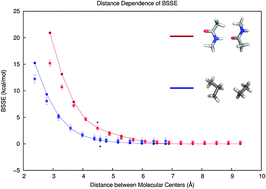
We describe a statistics-based model for the estimation of basis set superposition error (BSSE) for large biomolecular systems in which molecular fragment interactions are classified and analyzed with a linear model based on a bimolecular proximity descriptor. The models are trained independently for different classes of molecular interactions, quantum methods, and basis sets. The predicted fragment BSSE values, along with predicted uncertainties, are then propagated throughout the supermolecule to yield an overall estimate of BSSE and associated uncertainty. The method is described and demonstrated at the MP2/6-31G* and MP2/aug-cc-pVDZ levels of theory on a
- This article is part of the themed collection: Fragment and localized orbital methods in electronic structure theory

 Please wait while we load your content...
Please wait while we load your content...