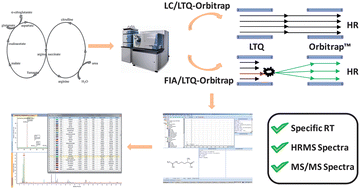
Metabolomics aims at detecting and semi-quantifying small molecular weight metabolites in biological samples in order to characterise the metabolic changes resulting from one or more given factors and/or to develop models based on diagnostic biomarker candidates. Nevertheless, whatever the objective of a metabolomic study, one critical step consists in the structural identification of mass spectrometric features revealed by statistical analysis and this remains a real challenge. Indeed, this requires both an understanding of the studied biological system, the correct use of various analytical information (retention time, molecular weight experimentally measured, isotopic golden rules, MS/MS fragment pattern interpretation…), or querying online databases. In gas chromatography-electro-ionisation (EI)-mass spectrometry, EI leads to a very reproducible fragmentation allowing establishment of universal EI mass spectra databases (for example, the NIST database – National Institute of Standards and Technology) and thus facilitates the identification step. Unfortunately, the situation is different when working with liquid chromatography-mass spectrometry (LC-MS) since atmospheric pressure ionisation exhibits high inter-instrument variability regarding fragmentation. Therefore, the constitution of LC-MS “in-house” spectral databases appears relevant in this context. The present study describes the procedure developed and applied to increment 133 and 130 metabolites in databanks dedicated to analyses performed with LC-HRMS in positive and negative electrospray ionisation, and the use of these databanks for annotating quickly untargeted metabolomics fingerprints. This study also describes the optimization of the parameters controlling the automatic processing in order to obtain a fast and reliable annotation of a maximum of organic compounds. This strategy was applied to bovine kidney samples collected from control animals or animals treated with steroid hormones. Thirty-eight compounds were identified successfully in the generated chemical phenotypes, among which five were found to be candidate markers of the administration of these anabolic agents, demonstrating the efficiency of the developed strategy to reveal and confirm metabolite structures according to the high-throughput objective expected from these integrative biological approaches.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...