Detecting and analyzing differentially activated pathways in brain regions of Alzheimer's disease patients†‡
Abstract
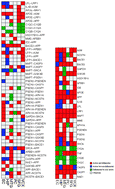
Alzheimer's disease (AD) generally results in neuronal loss due to protein dysfunction in various brain regions. Genome-wide data have provided new opportunities to analyze the underlying mechanisms of AD. Here, we present a novel network-based systems biology framework to identify and analyze differentially activated pathways by integrating human

 Please wait while we load your content...
Please wait while we load your content...