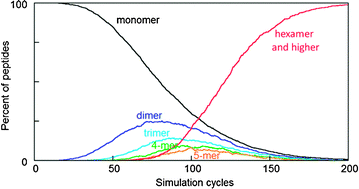
Severe conditions and lack of cure for many amyloid diseases make it highly desired to understand the underlying principles of formation of fibrillar aggregates (amyloid). Here, amyloid formation from peptides was studied using Monte Carlo simulations. Systems of 20, 50, 100, 200 or 500 hexapeptides were simulated. Association kinetics were modeled equal for fibrillar and other (inter- and intra-peptide) contacts and assumed to be faster the lower the effective contact order, which represents the distance in space. Attempts to form contacts were thus accepted with higher probability the lower the effective contact order, whereby formation of new contacts next to preexisting ones is favored by shorter physical separation. Kinetic discrimination was invoked by using two different life-times for formed contacts. Contacts within amyloid fibrils were assumed to have on average longer life-time than other contacts. We find that the model produces fibrillation kinetics with a distinct lag phase, and that the fibrillar contacts need to dissociate on average 5–20 times slower than all other contacts for the fibrillar structure to dominate at equilibrium. Analysis of the species distribution along the aggregation process shows that no other intermediate is ever more populated than the dimer. Instead of a single nucleation event there is a concomitant increase in average aggregate size over the whole system, and the occurrence of multiple parallel processes makes the process more reproducible the larger the simulated system. The sigmoidal shape of the aggregation curves arises from cooperativity among multiple interactions within each pair of peptides in a fibril. A governing factor is the increasing probability as the aggregation process proceeds of neighboring reinforcing contacts. The results explain the very strong bias towards cross β-sheet fibrils in which the possibilities for cooperativity among interactions involving neighboring residues and the repetitive use of optimal side-chain interactions are explored at maximum.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...