Abstract
To successfully perform biological experiments on bacteria in microfluidic devices, control of micron-scale
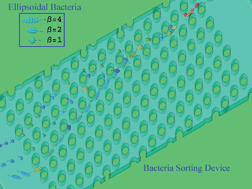
- This article is part of the themed collection: Emerging Investigators
* Corresponding authors
a
Department of Biomedical Engineering, Boston University, Boston, MA, USA
E-mail:
catherin@bu.edu
b Department of Mechanical Engineering, Boston University, Boston, MA, USA
To successfully perform biological experiments on bacteria in microfluidic devices, control of micron-scale

 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?
M. Kim and C. Klapperich, Lab Chip, 2010, 10, 2464 DOI: 10.1039/C003627G
To request permission to reproduce material from this article, please go to the Copyright Clearance Center request page.
If you are an author contributing to an RSC publication, you do not need to request permission provided correct acknowledgement is given.
If you are the author of this article, you do not need to request permission to reproduce figures and diagrams provided correct acknowledgement is given. If you want to reproduce the whole article in a third-party publication (excluding your thesis/dissertation for which permission is not required) please go to the Copyright Clearance Center request page.
Read more about how to correctly acknowledge RSC content.
 Fetching data from CrossRef.
Fetching data from CrossRef.
This may take some time to load.
Loading related content
