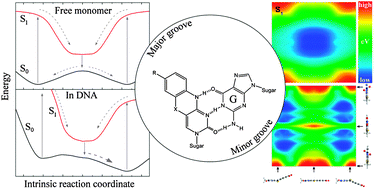
Fundamental insight into the unique fluorescence and nucleobase-mimicking properties of the fluorescent nucleobase analogues of the tC family is not only vital in explaining the behaviour of these probes in nucleic acid environments, but will also be profitable in the development of new and improved fluorescent base analogues. Here, temperature-dependent fluorescence quantum yield measurements are used to successfully separate and quantify the temperature-dependent and temperature-independent non-radiative excited-state decay processes of the three nucleobase analogues tC, tCO and tCnitro; all of which are derivatives of a phenothiazine or phenoxazine tricyclic framework. These results strongly suggest that the non-radiative decay process dominating the fast deactivation of tCnitro is an internal conversion of a different origin than the decay pathways of tC and tCO. tCnitro is reported to be fluorescent only in less dipolar solvents at room temperature, which is explained by an increase in excited-state dipole moment along the main non-radiative decay pathway, a suggestion that applies in the photophysical discussion of large polycyclic nitroaromatics in general. New insight into the ground and excited-state potential energy surfaces of the isolated tC bases is obtained by means of high level DFT and TDDFT calculations. The S0 potential energy surfaces of tC and tCnitro possess two global minima corresponding to geometries folded along the middle sulfur–nitrogen axis separated by an energy barrier of 0.05 eV as calculated at the B3LYP/6-311+G(2d,p) level. The ground-state potential energy surface of tCO is also predicted to be shallow along the bending coordinate but with an equilibrium geometry corresponding to the planar conformation of the tricyclic framework, which may explain some of the dissimilar properties of tC and tCO in various confined (biological) environments. The S1 equilibrium geometries of all three base analogues are predicted to be planar. These results are discussed in the context of the tC bases positioned in double-stranded DNA scenarios.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...