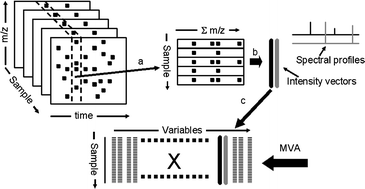
LC/MS is an analytical technique that, due to its high sensitivity, has become increasingly popular for the generation of metabolic signatures in biological samples and for the building of metabolic data bases. However, to be able to create robust and interpretable (transparent) multivariate models for the comparison of many samples, the data must fulfil certain specific criteria: (i) that each sample is characterized by the same number of variables, (ii) that each of these variables is represented across all observations, and (iii) that a variable in one sample has the same biological meaning or represents the same metabolite in all other samples. In addition, the obtained models must have the ability to make predictions of, e.g. related and independent samples characterized accordingly to the model samples. This method involves the construction of a representative data set, including automatic peak detection, alignment, setting of retention time windows, summing in the chromatographic dimension and data compression by means of alternating regression, where the relevant metabolic variation is retained for further modelling using multivariate analysis. This approach has the advantage of allowing the comparison of large numbers of samples based on their LC/MS metabolic profiles, but also of creating a means for the interpretation of the investigated biological system. This includes finding relevant systematic patterns among samples, identifying influential variables, verifying the findings in the raw data, and finally using the models for predictions. The presented strategy was here applied to a population study using urine samples from two cohorts, Shanxi (People’s Republic of China) and Honolulu (USA). The results showed that the evaluation of the extracted information data using partial least square discriminant analysis (PLS-DA) provided a robust, predictive and transparent model for the metabolic differences between the two populations. The presented findings suggest that this is a general approach for data handling, analysis, and evaluation of large metabolic LC/MS data sets.

 Please wait while we load your content...
Please wait while we load your content...