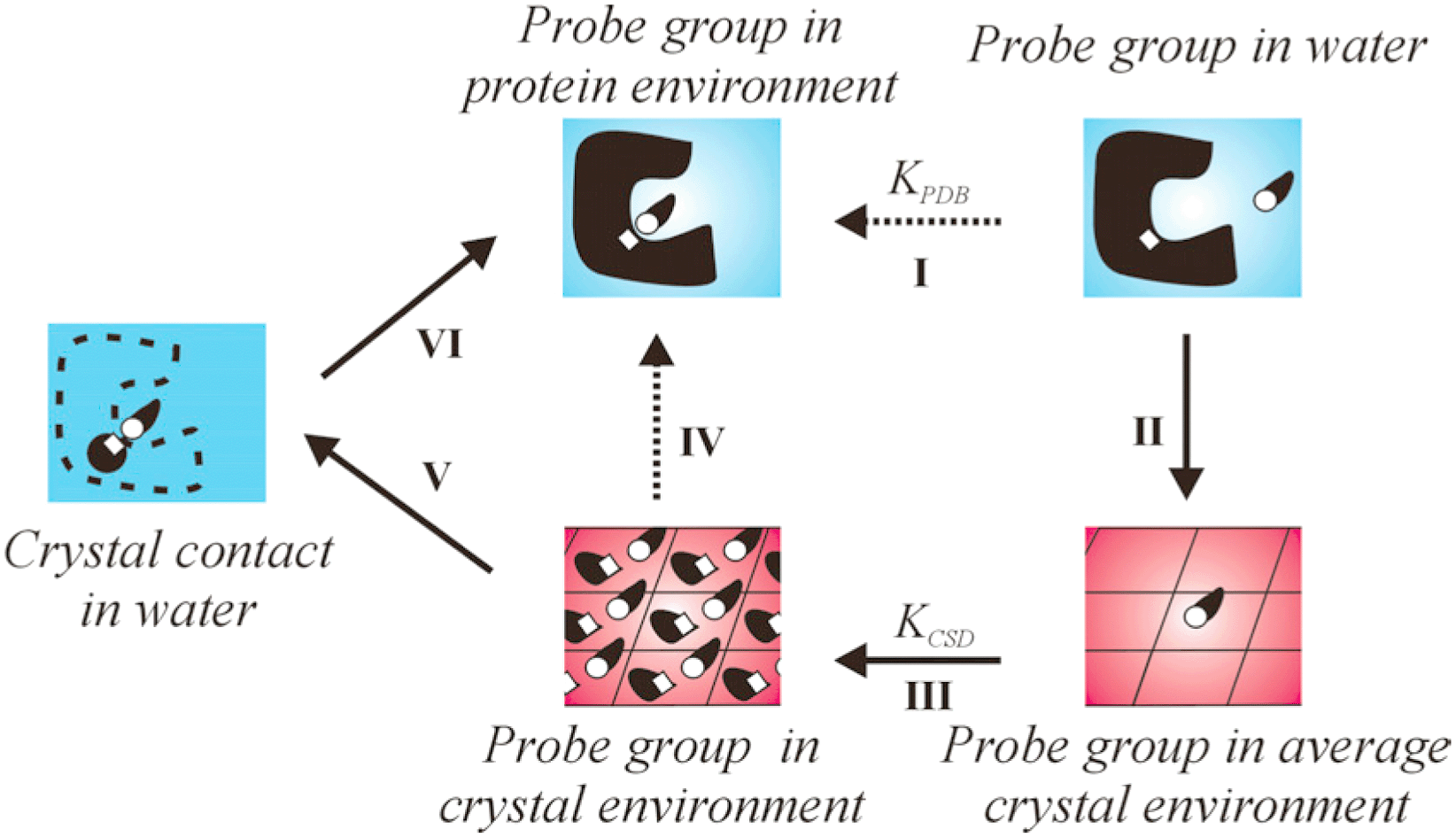
Knowledge-based methods are a good alternative to force-field-based methods for the analysis of sites of interaction in protein binding cavities. Both the Protein Data Bank (PDB) and the Cambridge Structural Database (CSD) offer a good amount of data on non-covalent interactions. Although different from protein-derived data, small-molecule crystal data from the CSD are worth looking at as they provide a much more abundant and diverse set of intermolecular contacts. CSD data, when properly corrected by use of octanol–water π values, can be used to predict the type of ligand chemical group most likely to occupy a given position within a protein binding site. Comparison with observed positions of ligand groups shows that the success rates of these predictions vary from 23% to 84%. Often, the group predicted to be most preferred at a given position is similar but not identical to the observed ligand group; if these are considered successes, prediction success rates range from 71% to 94%. Using PDB data, the corresponding rates are 16% to 79%, and 61% to 96%. Specificity of prediction of NH groups is somewhat better when using PDB interaction data, but results of prediction of hydrophobic groups seem worse than those obtained with CSD data.
We have analysed the importance of data selection by applying different filters to eliminate unwanted interactions from our knowledge-base. The presence of certain types of interactions can be undesirable if they are unrepresentative of biological situations (contact to solvent molecules in small-molecule crystal structures, secondary crystallographic contacts) or if they are likely to add noise to the data without conveying much new information (long-distance contacts, sparsely-populated data sets). The elimination of solvent contacts was found to have no effect on the prediction of ligand groups in our test set. Both secondary-contact filtering and noise filtering were found to have a clear beneficial effect on predictive ability.
You have access to this article
 Please wait while we load your content...
Something went wrong. Try again?
Please wait while we load your content...
Something went wrong. Try again?


 Please wait while we load your content...
Please wait while we load your content...