Proton and Cu(ii) binding to tren-based tris-macrocycles. Affinity towards nucleic acids and nuclease activity
Abstract
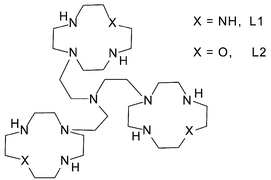
Proton binding by the two tren-based tris-macrocycles L1 and L2, composed, respectively, by three 1,4,7,10-tetrazacyclododecane ([12]aneN4) and three 1-oxa-4,7,10-triazacyclododecane ([12]aneN3O) macrocyclic moieties appended to a “tren” unit (tren = tris(2-aminoethyl)amine), has been analyzed by means of potentiometric and 1H and 13C NMR measurements in aqueous solutions. This study reveals that the ligands form highly charged polyammonium cations at neutral pH, containing six acidic protons equally distributed among the three macrocyclic units. A potentiometric and UV-vis spectrophotometric study shows that both ligands can form stable trinuclear Cu(II) complexes in a wide pH range. In the polynuclear complexes each metal is coordinated to a single macrocyclic unit. While the Cu(II) complexes with L1 do not show any tendency to form hydroxylated complexes, the mono-, di- and trinuclear L2 complexes give stable hydroxo-complexes, present in aqueous solutions from slightly acidic to alkaline pH values. Melting point studies indicate that the new tris-macrocyles and their Cu(II) complexes lead to stronger stabilization of double-stranded nucleic acids than those observed earlier with analogous ditopic macrocyclic ligands, again with preference for RNA-type polymers compared to DNA. The copper complexes promote cleavage of plasmid DNA and of bis-p-nitrophenyl phosphate (BNPP). Particular rate enhancements for BNPP with some complexes are attributed to the simultaneous action of three metal ions and partially to the formation of hydroxo complexes at neutral pH.

 Please wait while we load your content...
Please wait while we load your content...