Open Access Article
Open Access ArticleHarnessing a bis-electrophilic boronic acid lynchpin for azaborolo thiazolidine (ABT) grafting in cyclic peptides†
Basab Kanti
Das‡
,
Arnab
Chowdhury‡
 ,
Saurav
Chatterjee
,
Saurav
Chatterjee
 ,
Nitesh Mani
Tripathi
,
Nitesh Mani
Tripathi
 ,
Bibekananda
Pati
,
Soumit
Dutta
and
Anupam
Bandyopadhyay
,
Bibekananda
Pati
,
Soumit
Dutta
and
Anupam
Bandyopadhyay
 *
*
Biomimetic Peptide Engineering Laboratory, Department of Chemistry, Indian Institute of Technology Ropar, Rupnagar, Punjab 140001, India. E-mail: anupamba@iitrpr.ac.in
First published on 30th July 2024
Abstract
Chemical modifications of native peptides have significantly advanced modern drug discovery in recent decades. On this front, the installation of multitasking molecular grafts onto macrocyclic peptides offers numerous opportunities in biomedical applications. Here, we showcase a new class of borono-cyclic peptides featuring an azaborolo thiazolidine (ABT) graft, which can be readily assembled utilizing a bis-electrophilic boronic acid lynchpin while harnessing the inherent reactivity difference (>103 M−1 s−1) between the N-terminal cysteine and backbone cysteine for rapid and highly regioselective macrocyclization (∼1 h) under physiological conditions. The ABT-crosslinked peptides are fairly stable in endogenous environments, but can provide the linear diazaborine peptides via treatment with α-nucleophiles. This efficient peptide crosslinking protocol was further extended for regioselective bicyclizations and engineering of α-helical structures. Finally, ABT-grafted peptides were exploited in biorthogonal conjugation, leading to highly effective intracellular delivery of an apoptotic peptide (KLA) in cancer cells. The mechanism of action by which ABT-grafted KLA peptide induces apoptosis was also explored.
Introduction
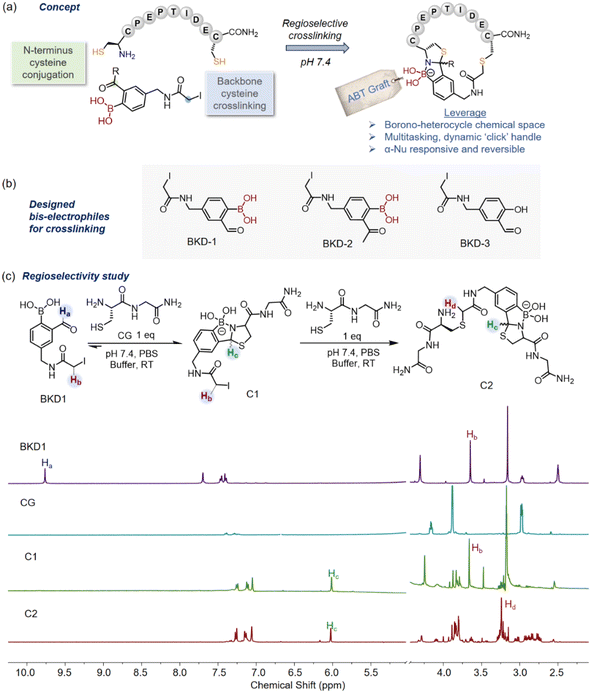
Peptides provide a unique three-dimensional chemical space between small molecules and large biologics, with enormous potential for drug development.1 However, as a consequence of their conformational freedom, most linear short peptides suffer from pharmacological liabilities, viz. proteolytic degradation, low binding affinity, and low target selectivity. To address these drawbacks, chemical modification and macrocyclization of bioactive peptides have proven advantageous and have emerged as a modern research hotspot for various disciplines.2 Peptide macrocyclization has enabled extensive chemical spacing, folding, and rigidification of peptide architectures, resulting in improved target binding, proteolytic and metabolic stability, and cell penetration.3 To date, various peptide macrocyclization methods are documented in the literature, each offering its own unique advantages. These include the formation of thioacetal,4–6 oxime,7 imine,8 and thioether9,10 linkages, as well as the deployment of click chemistry,11,12 olefin metathesis,13,14 and KAHA ligation15,16 regimens, along with many other approaches.17 A recent effort by Yudin and Chen's group has shown that the crosslinking of one or more aromatic side chains can achieve highly strained cyclophane-type macrocycles in peptide chains.18Contingent to the progress made in the chemoselective crosslinking of peptide backbones,19 the regioselective crosslinking of native peptides, especially cysteine-rich peptides, offers multiple opportunities in peptide engineering. For example, Brik's group adopted a de novo peptide cyclization strategy, forging two or three disulfide bonds in peptides or proteins in a chemo- and regioselective manner by judiciously utilizing the interplay between the orthogonal S-protecting groups on the native cysteine residues.20,21 Another study applied NHS-activated acrylamide for chemoselective amino-sulfhydryl crosslinking on native peptides.22 Recent efforts have also been directed towards the bis-electrophilic regioselective crosslinking of the N-terminus to backbone cysteines in phage-displayed peptide libraries.23–26 Nevertheless, in the context of peptide-based drug discovery, it is imperative to widen the scope of the regioselective crosslinking of cysteine-rich peptides for site-selective, exclusive macrocyclization27 towards the synthesis of functionally advanced molecular entities.28–30 Because the regioselective crosslinking of cysteine-rich peptides with bis-electrophiles is relatively less explored, there are ample opportunities to leverage the adjoining of versatile functional handles, such as heterocycles, click handles, covalent probes, and various synthetic scaffolds for wide prospects. For instance, derivatizing a variety of Lugdunin cyclopeptide31 analogs by the installation of thiazolidine-based molecular entities could lead to a potent antibacterial agent.
In our quest for engineered boronopeptides for biochemical applications,32–34 it was noted that borono-heterocycles are rarely seen in peptide frameworks and could provide an enormous range of novel biochemical applications. We conceptualized a regioselective peptide crosslinking method via the formation of azaborolo thiazolidine (ABT) grafts (Fig. 1a), in which the inherent Wulff-boronate35,36 biorthogonal handle and the thiazolidine37 moiety could be judiciously utilized to increase the chemical space. For instance, peptide frameworks modulated with heterocycles38 or boronic acid39 can facilitate cell penetration. In particular, boronic acid's versatile interaction modules (noncovalent and covalent)40 could provide diverse attractive prospects using this concept. It is also anticipated that the ABT graft could be released by specific α-nucleophiles to offer a linear peptide displaying diazaborine derivatives, which are of great interest for stimuli-responsive chemistry41 and the development of antibacterial agents.42 Towards this end, we now report a highly regioselective peptide macrocyclization using bis-electrophilic iodoacetamide tethered 2-formyl phenylboronic acid (2-FPBA) lynchpins, first through selective and quantitative engagement of the N-terminal cysteine to form azaborolo thiazolidine (ABT) grafts, followed by proximity-driven thioalkylation reactions with the backbone cysteines. This method further allows the facile construction of bicyclic peptides by establishing an additional disulfide bond in a regioselective manner. Finally, we demonstrate our efforts toward an engineered dual-functional peptide scaffold engaging ABT crosslinking, simultaneously folding a pro-apoptotic peptide into a secondary structure and delivering it into cancer cells by applying dynamic salicylhydroxamic boronate (SHAB)42 conjugation chemistry.
Results and discussion
Design of regioselective tandem bis-electrophile
The cysteine residue has featured in many chemoselective bioconjugation reactions due to its low interactome abundance and high nucleophilicity under physiological conditions.43–46 In this work, we envisaged regioselective crosslinking between two different types of cysteine found in native peptides, i.e., N-terminus cysteine and backbone cysteine, deploying bis-electrophilic lynchpins that were strategically functionalized with two kinetically distinctive electrophiles for selective orthogonal reactions (Fig. 1a and b). We combined a 2-FPBA moiety with the classical cysteine alkylating agent iodoacetamide (e.g., BKD1, Fig. 1b, ESI Section IIIa†), leveraging the superior reactivity of N-terminal cysteines with 2-FPBA (>103 M−1 s−1)47vis-a-vis iodoacetamide-mediated S-alkylation to ensure orthogonal cross reactivity. The native peptides were expected to first react with BKD-1 preferentially via the N-terminus cysteine to form a thermodynamically stable azaborolo thiazolidine (ABT) conjugate and subsequently engage in proximity-driven S-alkylation with backbone cysteine, leading to macrocyclization. The macrocyclic ABT conjugates could be further edited as a dynamic ‘click’ handle42 while benefitting from boronic acid (BA) fusion with the thiazolidine heterocycle37 for diverse biological submissions. As controls, we also prepared two other bis-electrophiles, BKD2 and BKD3, based on 2-acetyl phenylboronic acid (2-APBA) and 2-hydroxy benzaldehyde (2-HB) (Fig. 1b). While 2-APBA is known to combine rapidly with N-terminus cysteine at physiological pH, its ABT conjugate is thermodynamically less stable compared to 2-FPBA.42 The 2-HB derivatives are also known to produce thiazolidine derivatives with N-terminal cysteine but with sluggish kinetics even at a slightly acidic pH.47The regioselectivity of the designed bis-electrophiles BKD1-3 towards cysteine bioconjugation was probed through 1H-NMR experiments using a cysteine dipeptide (CG) in PBS buffer (pH 7.4) (Fig. 1c). An equimolar mixture of CG and BKD1 (∼1 mM each) resulted in a rapid disappearance of the formyl proton at 9.85 ppm with the concomitant appearance of two diastereomeric protons of the ABT conjugate (C1) at around 6.0–6.2 ppm. ABT conjugate formation was further confirmed by the upfield shifting of the aromatic protons, accompanied by enhanced splitting. The addition of an extra equivalent of CG to this mixture caused an upfield shift of the side-chain iodoacetamide methylene protons from 3.68 ppm to 3.23 ppm, indicating side-chain S-alkylation to form the bis-adduct C2 (Fig. 1c). On the other hand, the addition of ∼1.0 equivalent of CG to BKD2 only caused an upfield shift of the methylene protons of BKD2, indicating the occurrence of the promiscuous S-alkylation of the iodoacetamide side chain as the primary reaction (Fig. S1†), perhaps due to the reversible nature of ABT conjugation with the more sterically hindered 2-APBA probe. Furthermore, treatment of salicylaldehyde-derived probe BKD3 with CG only indicated the side-chain S-alkylated product with no thiazolidine formation, even with 2.0 equivalents of CG. (Fig. S2†). Based on these findings, the 2-FPBA-derived BKD-1 emerged as the optimum bis-electrophile for our crosslinking purposes.
Investigation of regioselective peptide crosslinking
The crosslinking ability of BKD-1 was examined using the model peptide CIITRPC. After incubation of equimolar concentrations (∼100 μM) of BKD-1 and CIITRPC at pH 7.4 for only 30 min, a prominent peak corresponding to the crosslinked peptide P1 appeared in the LC-MS spectrum (Fig. 2a, ESI Section V†), confirming our hypothesis that the 2-FPBA based bis-electrophilic lynchpins were highly effective cysteine crosslinking scaffolds for the regioselective macrocyclization of native peptides. In contrast, as expected, treating BKD-2 with the peptide CIITRPC yielded two major peaks in the LC, indicating the formation of two side-chain cysteine-alkylated products, as confirmed by mass analysis (ESI Fig. S3†). Notably, the crosslinked peptide P1 generated a mixture of two diastereomers around the thiazolidine heterocycle. The presence of diastereomeric thiazolidines in the natural cyclopeptide lugdunin,31 an antibiotic, served as an additional impetus to pursue our ABT crosslinking campaign.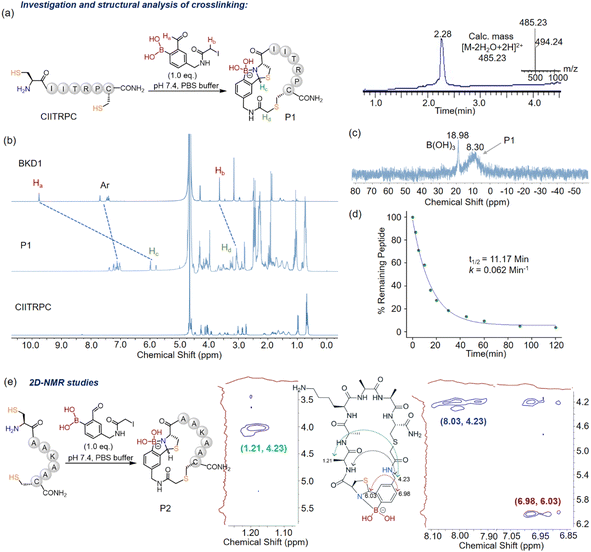 | ||
Fig. 2 (a) Scheme of the peptide crosslinking with BKD1 resulting in the macrocyclization of the model peptide CIITRPC. The LC-MS profile of the reaction mixture containing CIITRPC and BKD1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) shows the expected product as the major component. (b) Investigation of crosslinking of the model peptide using 1H-NMR in (1 1) shows the expected product as the major component. (b) Investigation of crosslinking of the model peptide using 1H-NMR in (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) D2O 4) D2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS (7.4). Three major changes in the 1H-NMR chemical shifts of the peaks of BKD1 (viz. formyl, aromatic, and methylene iodide) demonstrate the regioselective crosslinking. (c) 11B-NMR spectrum of P1, which clearly establishes the N–B connection in the azaborolo thiazolidine (ABT) graft. (d) Reaction kinetics of crosslinking at pH 7.4 monitored through LC-MS over time by quenching with 1 M HCl (for details, see ESI Section VI†); the peak area of the leftover CIITRPC in the reaction mixture at certain time intervals was calculated to monitor the reaction progress. (e) Assigned NOEs in the ROESY spectra, demonstrating the regioselective crosslinking in P2. The colored arrows in the P2 chemical structure represent the primary NOEs observed, which correlate with 2D-NMR data shown in brackets. PBS (7.4). Three major changes in the 1H-NMR chemical shifts of the peaks of BKD1 (viz. formyl, aromatic, and methylene iodide) demonstrate the regioselective crosslinking. (c) 11B-NMR spectrum of P1, which clearly establishes the N–B connection in the azaborolo thiazolidine (ABT) graft. (d) Reaction kinetics of crosslinking at pH 7.4 monitored through LC-MS over time by quenching with 1 M HCl (for details, see ESI Section VI†); the peak area of the leftover CIITRPC in the reaction mixture at certain time intervals was calculated to monitor the reaction progress. (e) Assigned NOEs in the ROESY spectra, demonstrating the regioselective crosslinking in P2. The colored arrows in the P2 chemical structure represent the primary NOEs observed, which correlate with 2D-NMR data shown in brackets. | ||
Encouraged by the prospects of BKD-1 as an efficient cysteine crosslinking module, we undertook detailed 1H- and 11B-NMR studies of its reaction with CIITRPC in PBS buffer. Upon crosslinking with CIITRPC, BKD-1 exhibited distinct proton chemical shifts due to the formation of the ABT conjugate and S-alkylation of the iodoacetamide unit (Fig. 2b), a pattern which is similar to the signature shown by C2 (cf.Fig. 1c). In the 11B-NMR spectrum, the ABT unit in the crosslinked peptide P1 showed a chemical shift at 8.30 ppm (Fig. 2c), indicating the exclusive population of N–B connected heterocycles.47 BKD1 alone shows a chemical shift at ∼30 ppm in an 11B-NMR spectrum (ESI Section VIII and Fig. S7†). To explore the kinetics of the cyclization process, we performed LC-MS analysis of the reaction between CIITRPC and BKD1 (∼100 μM each) over time and calculated the half-life (Fig. 2d, ESI Section VI and Fig. S4†). The obtained k value aligns with the proximity-driven S-alkylation reaction (first-order kinetics), providing additional support to our hypothesis. The initial ABT formation occurred rapidly (>103 M−1 s−1)47 within a few seconds at 100 μM so that the subsequent S-alkylation step turned into the rate-determining (RD) step (t1/2 ∼ 11 min), governing the overall cyclization kinetics. However, the kinetics could differ with varying the length of the peptide sequence.
The veracity of the macrocyclization was further proved using various 2D NMR experiments (COSY, TOCSY, and ROESY) to examine the crosslinking of the CAAKAAC peptide with BKD-1 to produce P2 in DMSO-d6 (Fig. 2e, ESI Section VII and Fig. S5, S6†). The ROESY experiment elicited significant inter-residue cross-peaks, such as Ala–CH3–β(H) ↔ Benz–CH2 (strong), Ala–NH ↔ Benz–CH2 (medium), ABT–C–H ↔ Ar–H and ABT–C–H ↔ Benz–CH2 (medium), supporting the presence of a stable macrocyclic structure in the BKD-1 crosslinked peptide (Fig. 2e). Additionally, clean conversions were observed within the range of 1 mM substrate concentration (ESI Section IX and Fig. S8†), whereas higher probe concentrations (>1 mM) led to a loss in reaction orthogonality on the cysteine centers.
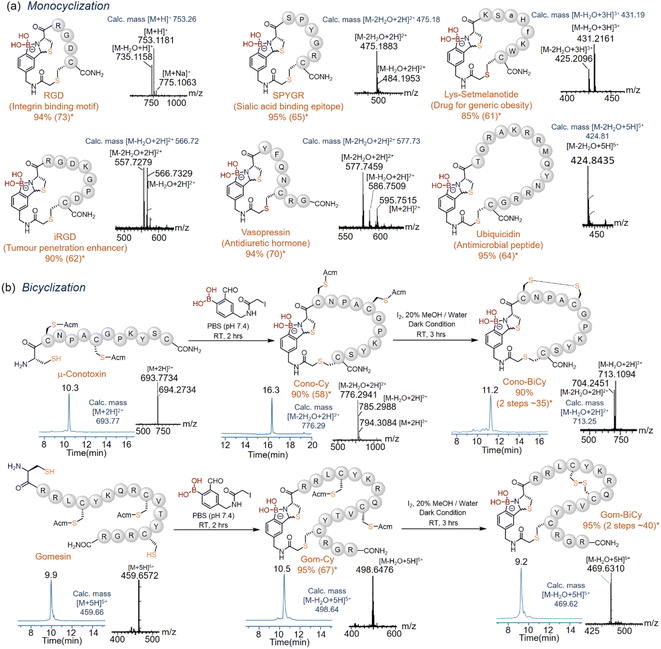
Substrate scope for monocyclization and bicyclization
The scope of this novel crosslinking methodology with BKD-1 was explored with various biologically active peptide sequences (RGD (shortest) and gomesin (largest)). Most peptides showed >90% regioselective crosslinked product conversion in PBS (pH 7.4) within 60 min, independent of the length and amino acid sequences (Fig. 3a), as was evident from the recorded HPLC, mass (ESI Section X and Fig. S9–S14†) and 1H-NMR data (ESI Section XI and Fig. S15–S17†). The crosslinked thiazolidine diastereomers did not separate in the routinely used HPLC gradient, but in some cases, a hump or a broad peak was observed near the crosslinked peptide peak due to the protonation of the N-atom in the ABT motif in acidic HPLC buffer. All peptides were isolated through HPLC purifications with a good yield (Fig. 3a). Notably, this crosslinking could be executed at a low micromolar concentration of BKD1 in a quantitative manner due to a selective, rapid, and highly productive association between 2-FPBA and N-terminal cysteine,48 based on the advantage of the designed lynchpin, which avoids the side reactions of using the over-alkylating (iodoacetamide) agent.49A second disulfide ring could also be appended to the crosslinked ABT peptides via strategic combination with the iodine-mediated disulfide crosslinking of Acm-protected cysteine pairs.50 Towards this end, native peptides containing an additional pair of Acm-protected cysteines, viz. those derived from conotoxin and gomensin (Fig. 3b), were first subjected to BKD-1 coupling, resulting in rapid crosslinking between the free N-terminus and a backbone cysteine to produce Cono-Cy and Gom-Cy in near-quantitative conversions. The latter, upon iodine-mediated regioselective disulfide bridging between the Acm-protected cysteine pairs, then appended the second ring34 to furnish the bicyclic peptides Cono-BiCy and Gom-BiCy, respectively (ESI Section XII and Fig. S18, S19†). This simplified approach for regioselective bicyclization in solution-phase peptide chemistry holds great promise for the synthesis of bioactive bicyclic peptides with an enhanced chemical space and desirable biological properties.51
Stability and reversibility of crosslinked peptides
Due to the labile nature of the thiazolidine ring at acidic pH values,47 the solution stability of the cyclic ABT peptides was also investigated, e.g., with P1via1H-NMR (Fig. 4a). The cyclic peptide P1 was found to be stable in a PBS buffer (pH = 7.4) over 15 days, as shown in the LC traces beyond that time (ESI Section XIV and Fig. S21†). With lowering of the pH, slight downfield shifts and broadening of the aromatic protons and thiazolidine ring were observed, perhaps due to dynamic protonation and deprotonation of the N-atom in the ABT graft in acidic media (Fig. 4a, ESI Section XIII†). However, complete dissociation of the thiazolidine ring with the appearance of an aldehyde peak corresponding to the 2-FPBA moiety was never observed, demonstrating the high stability of P1 under a wide range of pH values. Contrary to the rapid oxidation of phenylboronic acid by H2O2 under physiological conditions,52 cyclic peptide P1 showed remarkable stability in ∼150 μM H2O2 even after 6 h (Fig. 4c, ESI Section XV†) due to strong coordination between N and B atoms in the ABT motif. A maximum concentration of ∼100 μM can be found in the case of malignant tumor cells,53 and ABT-crosslinked peptide showed notable compatibility in the most abundant reactive oxygen species (ROS) environment. Interestingly, we found that ABT-crosslinked ubiquicidine (UBI 29–41, Fig. 3a) peptide exhibited a half-life 10 times that of the linear precursor in human serum (∼20% diluted in PBS, Fig. 4b). UBI, an antimicrobial peptide, was modified with N- and C-terminus Cys for ABT crosslinking. The Cys residues of the linear analog (linear UBI) were alkylated with iodoacetamide to restrict oxidative cyclization in the physiological buffer system.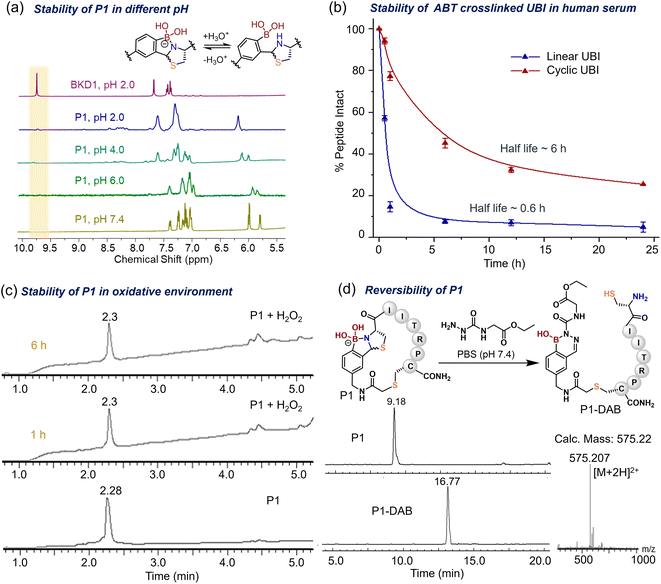 | ||
| Fig. 4 Stability studies of crosslinked peptides. (a) 1H-NMR profile of P1 at different pH values. The aromatic region is shown for better resolution and compared with the BKD1 profile. (b) Serum (∼20% in PBS 7.4) stability of ABT-crosslinked ubiquicidine (UBI 29–41) peptide compared with that of the linear UBI 29–41 peptide. The crosslinked UBI showed a 10-fold increase in stability. (c) LC-MS profile of P1 (∼100 μM) treated with ∼150 μM hydrogen peroxide in PBS (pH 7.4) for up to 6 h, demonstrating its stability (Method 1 Section Ic in ESI†). (d) Linearization of P1 investigated using HPLC and mass in the presence of ∼100 μM semicarbazide derivative in PBS (pH 7.4) for 20 min (Method 3 Section Ic in ESI†). | ||
Interestingly, the ABT peptide P1 could be linearized with an equimolar amount of the biocompatible nucleophile semicarbazide54 in PBS (7.4) due to the formation of a highly stable diazaborine conjugate, as confirmed using LC-MS. The peptide (P1) linearization via diazaborine formation was further confirmed using 1H-NMR after treatment with phenylhydrazine (ESI Section XVI and Fig. S23†), demonstrating that this crosslinking strategy responds to certain chemical stimuli such as semicarbazide and hydrazine. However, the ABT unit appeared to be stable against exchange from free cysteine, as demonstrated by time-dependent NMR analysis of the reaction between P1 (∼1 mM) and free cysteine (∼5 mM) in (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) D2O
4) D2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS (7.4) for 10 hours (ESI Section XVII and Fig. S24†). The above studies clearly demonstrated that the ABT-crosslinked peptides show high chemical stability against endogenous stimuli such as acidic pH, ROS, and cysteamine-group-containing molecules.
PBS (7.4) for 10 hours (ESI Section XVII and Fig. S24†). The above studies clearly demonstrated that the ABT-crosslinked peptides show high chemical stability against endogenous stimuli such as acidic pH, ROS, and cysteamine-group-containing molecules.
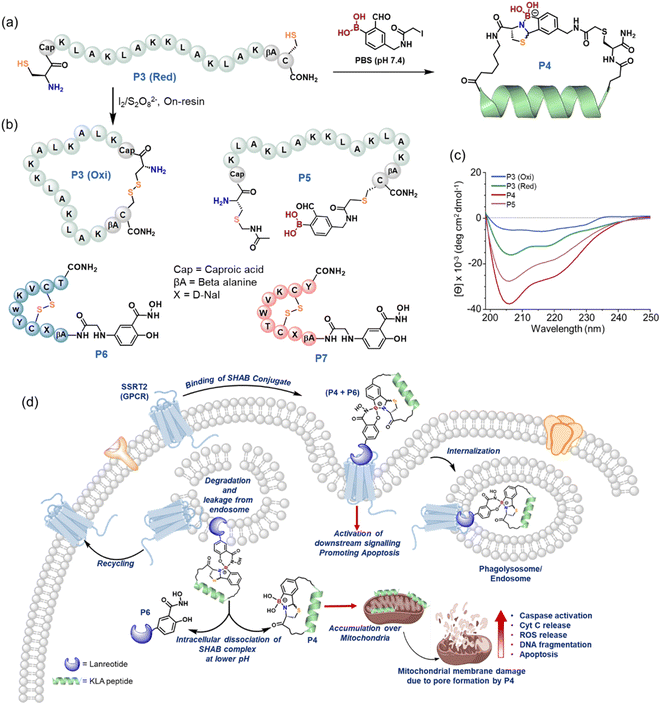
Crosslinking for folding and intracellular delivery
To augment the significance of the boronic acid lynchpin, we postulated that the method would permit the spontaneous folding of a biologically active peptide into a favorable secondary structure. Further, to facilitate the intracellular delivery of the peptide, the boronic acid moiety can be used to conjugate a drug that is reported to selectively bind to cell surface receptors. In order to investigate the proposal, KLA peptide55,56 was remodeled in silico (ESI Section XVIII†) with appropriate spacers and cysteine, which was expected to enhance the total helical content upon ABT crosslinking (Fig. 5a). The KLA peptide is derived from melittin and exhibits a strong affinity for negatively charged phospholipid membranes.55,56 Its helical conformation induces pore formation on the surfaces of mitochondrial membranes. Consequently, the release of cytochrome-c into the cytosol is triggered, activating apoptotic or necrotic cell death pathways,57–59 highlighting its potency in combating cancer. Towards this end, the designed KLA linear precursor (P3 (Red)) was crosslinked with BKD1 to yield P4 (ESI Section XIX and Fig. S27†) and oxidized to prepare a disulfide-crosslinked KLA control (P3 (Oxi)). Circular dichroism (CD) analyses illustrated a >5-fold enhancement of helical content in KLA upon ABT crosslinking (P4) compared to P3 (Oxi) (Fig. 5c, ESI Section XX†), which is in agreement with computational estimations (Fig. S25†). In a control CD experiment, an S-alkylated linear KLA (P5) peptide having a 2-FPBA moiety (Fig. 5b) demonstrated a ∼3.6-fold increase in peptide folding compared to P3 (Oxi). We believe that the spontaneous and random cyclization via iminoboronate formation60 between 2-FPBA and the lysine side chains in P5 resulted in these improvements in helicity, which verifies that the appropriate crosslinking of the KLA peptide is essential for better helical folding. Overall, the strategic ABT crosslinking of KLA peptide enabled helical folding to be induced to a greater extent.We further sought to explore whether P4 (ABT-crosslinked) exhibits better apoptotic activity than P5 (linear). Notably, the endocytosis of KLA peptide is limited to eukaryotic cells,61–63 and the cellular entry can be enhanced by conjugating KLA with targeting motifs such as hyaluronic acid64,65 or polyarginine.65 However, our methodology could address this limitation via the appended boronic acid moiety in crosslinked KLA (P4). We postulated that a somatostatin peptide analog functionalized with the salicylhydroxamic (SHA) moiety could readily conjugate with the crosslinked KLA peptide (P4) via salicylhydroxamic boronate (SHAB) formation, and that the combined formulate could selectively bind to the overexpressed somatostatin receptor in cancer cells, similar to an antibody–drug conjugate. Further, the conjugate would be endocytosed to release P4, triggered by the lower pH in cytosol, which would subsequently induce apoptosis (Fig. 5d). In addition, the combined antiproliferative activity of the somatostatin analog and crosslinked KLA could represent a better approach to combating cancer.66 In order to test this hypothesis, lanreotide (LAN),67 a synthetic analog of somatostatin that binds to SSR2 selectively, was engineered with an SHA moiety at the N-terminus (P6, Fig. 5b, ESI Section XXI and Fig. S28†). A scrambled LAN peptide adjoined to SHA (P7) and P5 was considered as controls. The formation of SHAB conjugates between P4 and P6 was confirmed using mass spectrometry (ESI Section XXIII and Fig. S29†) before assessing its antiproliferative activity.
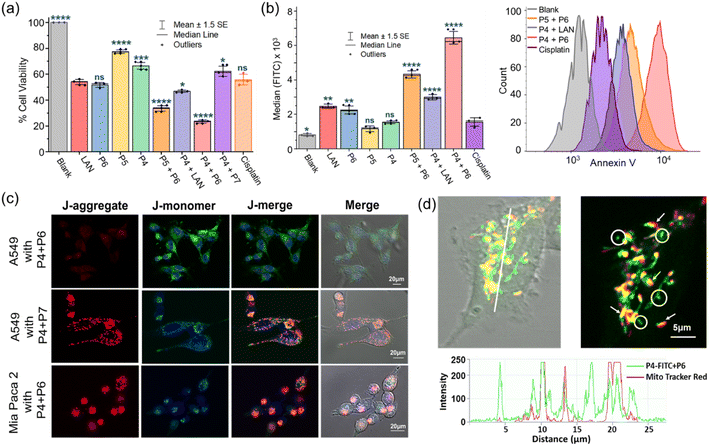
Moving forward, we performed MTT assays on the A549 cell line to evaluate our hypothesis of combined therapy. Individual peptides (P4–6, ∼20 μM) and three combinations (P4 + P6, P5 + P6, and P4 + P7) prepared by mixing equimolar amounts of each (∼20 μM in culture media) were used in this study. Cisplatin (∼50 μM) and LAN (∼20 μM) were included as positive controls (ESI Section XXIV and Fig. S30a†). After a 12 hour incubation period, we observed that the combination of P4 + P6 exhibited the highest cytotoxicity (∼24% cell viability), which is >2-fold higher than that of the individual peptides (Fig. 6a). Although P4 and P5 are both anticipated to cross the extracellular membrane with the assistance of P6via SHAB conjugation, P5 + P6 showed a slightly reduced cytotoxic response compared to P4 + P6 (∼34% vs. ∼24% cell viability), perhaps due to the lower helical content and lower enzymatic stability of P5 than P4. As expected, the individual treatment of A549 with P5 or P4 exhibited the least cytotoxicity owing to their poor cell permeability. P4 provided better results than P5 to some extent; it probably existed in a compact ABT-crosslinked structure. When P4 was combined with scrambled LAN (P7), the cell apoptosis was compromised ∼2.5-fold compared to that of (P4 + P6), proving the SSR2-assisted delivery of P4 (Fig. 6a). The SSR2-mediated delivery was further justified the very low antiproliferative activity of P4 + P6 in cell lines lacking SSR2 receptors, such as Mia PaCa-2 (Fig. S30b†). The data unambiguously demonstrate the importance of the suitable crosslinking of KLA and the conjugation of KLA with suitable receptor-targeting peptides to create an effective biological response. Further, the control experiment of combined P4 + LAN treatment exhibited a greatly reduced response (∼47% cell viability) compared to the two combinations of P4 + P6 and P5 + P6, unambiguously demonstrating the importance of boronic-acid-mediated SHAB conjugation in intracellular peptide delivery. The selective apoptotic activity of P4 + P6 in normal cells (HEK329) and cancer cells (A549) was compared, demonstrating that the conceptualized combination therapy is ∼2.5-fold more selective toward cancer cells (Fig. S30b†).
We were keen to quantitatively assess the apoptotic activity of combined therapy events to strengthen our hypothesis. Cell apoptosis was monitored in flow cytometry by labeling externalized phosphatidylserine with annexin V-FITC (ESI Section XXV and Fig. S31†). The fluorescence readout (Fig. 6b) demonstrated a consistent outcome ascribed to the MTT assay, in which P4 + P6 combinedly displayed ∼1.45 and ∼2.1-fold higher fluorescence median value intensities than the apoptotic activity of P5 + P6 and P4 + LAN, respectively. Notably, the SHAB conjugate (P4 + P6) showed >2.5-fold higher antiproliferative activity compared to any individual component. Together, these findings validate the potential of combination therapy, demonstrate successful intracellular delivery, and highlight the uniqueness of this crosslinking strategy.
Mechanism of action
We set out to reinvestigate the mechanism of action of the apoptotic activity shown by P4. KLA peptides are known to damage mitochondrial membranes, which results in depolarization of the mitochondrial membrane with a drop in potential (Δψm).68 This work examined the mitochondria-regulated apoptosis pathway using fluorescence probe JC-1 under a laser scanning confocal microscope (LSCM). It has been reported that the green monomer of JC-1 enters the cytoplasm and forms red fluorescent aggregates in normal mitochondria, namely red J-aggregates, which revert to the monomer stage (green) upon depolarization of the mitochondrial membranes.69 In view of this mechanism, we noticed the disappearance of red fluorescence (normal mitochondria) and an increase in green fluorescence (monomer in the cytoplasm) in A549 cells when incubated with P4 + P6 under LSCM (Fig. 6c, top row). This phenomenon could only be ascribed to serious damage of the mitochondrial membrane of A549 cells by P4via SSR2-mediated entry, leading to a relatively low Δψm. In contrast, there is no obvious change in Δψm for Mia Paca 2 cells due to the lack of SSRT2-mediated entry of P4 (Fig. 6c, bottom row). A similar status of A549 was detected for treatment with P4 + P7 (Fig. 6c, middle row), indicating that the LAN sequence is essential for SSRT2-mediated cellular entry. In the case of normal epithelial cell line HEK293, a small amount of P4 + P6 entry was revealed, because normal cells also express the SSRT2 receptor, but not to the extent of cancerous cells (Fig. S32†); these results were consistent with those of the MTT assays. We noticed that treatment of A549 with P4 alone damaged neither the plasma membrane nor the mitochondrial membrane under LSCM (Fig. S32†). Finally, treatment with FITC-labeled P4 (P4-FITC, Fig. S26†) for 4 hours, followed by Mito Tracker red, revealed the overlapped regions of green and red fluorescence under LSCM, thus unambiguously demonstrating the P4 selectivity toward mitochondria (Fig. 6d, shown by arrows). We assumed that the green aggregates (Fig. 6d, indicated by circles) that do not overlap with the Mito Tracker are phagolysosomes that slowly released P4 to reach the mitochondria. The fluorescence signals (Fig. 6d, bottom intensity graph) that are grounded on the white line perhaps indicate such phenomena.Conclusion
The report summarizes a new regioselective crosslinking regime, resulting in a new class of borono-cyclopeptides for biochemical exploitation by deploying a bis-electrophilic lynchpin. The designed tandem bis-electrophile, which is constructed by uniting ortho carbonyl phenylboronic acid and iodoacetamide, manifested rapid crosslinking between the N-terminus and backbone cysteine at physiological pH at micromolar concentration. This technology is high-yielding and operationally simple, irrespective of the amino acid residues or chain lengths of the peptides. Interestingly, crosslinked peptides exhibited stability at physiological and acidic pH and under a physiologically significant ROS stimulus (H2O2). The crosslinked peptide could be linearized upon suitable applications of hydrazine stimuli by releasing an important diazaborine pharmacophore, which proved the superiority of this crosslinking method. The method clearly achieves regioselective bicyclization, as evidenced by conotoxin and gomesin construction, which could create multiple valuable opportunities in drug discovery platforms. The ABT graft that achieved the cyclopeptide frameworks showed further utility in folding, drug delivery, and combined cancer therapy. We showed that the ABT-graft-induced folded pro-apoptotic peptide could be delivered to mitochondria with the assistance of a somatostatin analog via dynamic SHAB chemistry to achieve improved retardation of cancer growth. In a nutshell, our investigation advances two important goals of chemical biology, including providing a solution to the synthetic challenge of late-stage modification of peptide cyclization in therapeutic conditions and expanding the nexus of possibilities of cyclic peptides for modern drug discovery. We speculate that our contribution will pave the way for many directions in peptide therapeutics.Data availability
The data supporting this article have been included in the ESI.†Author contributions
AB conceptualized and supervised the project. BKD performed all crosslinking chemistry and analysis with the assistance of SC, BNP, and SD. All biological experiments were performed by AC. NMT helped in computational model design and MD simulation. The manuscript was written by the contribution of all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully welcome the financial assistance provided by IIT Ropar through the ISIRD grant and the core research grant (CRG/2022/007757) from SERB, India. BKD thanks IIT Ropar for the postdoctoral fellowship. SC and AC express gratitude to IIT Ropar for providing a PhD fellowship. NMT expresses gratitude to DST for giving a fellowship. We also appreciate the IIT Ropar central research facility for the HRMS and NMR facilities and the department LCMS facility (SR/FST/CS-I/2018/55). The IIT Ropar HPC facility was used for all computations. We are grateful to Kashiv Biosciences in India for their assistance.Notes and references
- A. Capecchi and J. L. Reymond, Med. Drug Discovery, 2021, 9, 100081 CrossRef CAS.
- C. Bechtler and C. Lamers, RSC Med. Chem., 2021, 12, 1325–1351 RSC.
- A. Boto, C. C. González, D. Hernández, I. Romero-Estudillo and C. J. Saavedra, Org. Chem. Front., 2021, 8, 6720–6759 RSC.
- C. M. B. K. Kourra and N. Cramer, Chem. Sci., 2016, 7, 7007–7012 RSC.
- S. Lindman, G. Lindeberg, P.-A. Frändberg, F. Nyberg, A. Karlén and A. Hallberg, Bioorg. Med. Chem., 2003, 11, 2947–2954 CrossRef CAS PubMed.
- M. Ueki, T. Ikeo, K. Hokari, K. Nakamura, A. Saeki and H. Komatsu, Bull. Chem. Soc. Jpn., 1999, 72, 829–838 CrossRef CAS.
- L. R. Malins, J. N. deGruyter, K. J. Robbins, P. M. Scola, M. D. Eastgate, M. R. Ghadiri and P. S. Baran, J. Am. Chem. Soc., 2017, 139, 5233–5241 CrossRef CAS PubMed.
- B. Li, Z. Wan, H. Zheng, S. Cai, H.-W. Tian, H. Tang, X. Chu, G. He, D.-S. Guo, X.-S. Xue and G. Chen, J. Am. Chem. Soc., 2022, 144, 10080–10090 CrossRef CAS PubMed.
- Y. Goto, A. Ohta, Y. Sako, Y. Yamagishi, H. Murakami and H. Suga, ACS Chem. Biol., 2008, 3, 120–129 CrossRef CAS PubMed.
- N. Bionda, A. L. Cryan and R. Fasan, ACS Chem. Biol., 2014, 9, 2008–2013 CrossRef CAS PubMed.
- W. S. Horne, C. A. Olsen, J. M. Beierle, A. Montero and M. R. Ghadiri, Angew. Chem., Int. Ed., 2009, 48, 4718–4724 CrossRef CAS PubMed.
- M. G. Wuo, A. B. Mahon and P. S. Arora, J. Am. Chem. Soc., 2015, 137, 11618–11621 CrossRef CAS PubMed.
- Y.-W. Kim, T. N. Grossmann and G. L. Verdine, Nat. Protoc., 2011, 6, 761–771 CrossRef CAS PubMed.
- H. E. Blackwell, J. D. Sadowsky, R. J. Howard, J. N. Sampson, J. A. Chao, W. E. Steinmetz, D. J. O'Leary and R. H. Grubbs, J. Org. Chem., 2001, 66, 5291–5302 CrossRef CAS PubMed.
- T. Fukuzumi, L. Ju and J. W. Bode, Org. Biomol. Chem., 2012, 10, 5837–5844 RSC.
- J. W. Bode, R. M. Fox and K. D. Baucom, Angew. Chem., Int. Ed., 2006, 45, 1248–1252 CrossRef CAS PubMed.
- H. C. Hayes, L. Y. P. Luk and Y. H. Tsai, Org. Biomol. Chem., 2021, 19, 3983–4001 RSC.
- P. Yang, M. J. Širvinskas, B. Li, N. W. Heller, H. Rong, G. He, A. K. Yudin and G. Chen, J. Am. Chem. Soc., 2023, 145, 13968–13978 CrossRef CAS PubMed.
- C. Bechtler and C. Lamers, RSC Med. Chem., 2021, 12, 1325–1351 RSC.
- S. Laps, F. Atamleh, G. Kamnesky, S. Uzi, M. M. Meijler and A. Brik, Angew. Chem., Int. Ed., 2021, 60, 24137–24143 CrossRef CAS PubMed.
- S. Laps, F. Atamleh, G. Kamnesky, H. Sun and A. Brik, Nat. Commun., 2021, 12, 870 CrossRef CAS PubMed.
- M. J. S. A. Silva, H. Faustino, J. A. S. Coelho, M. V Pinto, A. Fernandes, I. Compañón, F. Corzana, G. Gasser and P. M. P. Gois, Angew. Chem., Int. Ed., 2021, 60, 10850–10857 CrossRef CAS PubMed.
- T. R. Oppewal, I. D. Jansen, J. Hekelaar and C. Mayer, J. Am. Chem. Soc., 2022, 144, 3644–3652 CrossRef CAS PubMed.
- Y. Wu, C. Li, S. Fan, Y. Zhao and Y. Zhao, Bioconjugate Chem., 2021, 32, 2065–2072 CrossRef CAS PubMed.
- X. Zheng, Z. Li, W. Gao, X. Meng, X. Li, L. Y. P. Luk, Y. Zhao, Y. H. Tsai and C. Wu, J. Am. Chem. Soc., 2020, 142, 5097–5103 CrossRef CAS PubMed.
- J. T. Hampton, T. J. Lalonde, J. M. Tharp, Y. Kurra, Y. R. Alugubelli, C. M. Roundy, G. L. Hamer, S. Xu and W. R. Liu, ACS Chem. Biol., 2022, 17, 2911–2922 CrossRef CAS PubMed.
- R. Wills, V. Adebomi and M. Raj, ChemBioChem, 2021, 22, 52–62 CrossRef CAS PubMed.
- I. V. Smolyar, A. K. Yudin and V. G. Nenajdenko, Chem. Rev., 2019, 119, 10032–10240 CrossRef CAS PubMed.
- B. J. Wang and M. P. Groziak, Adv. Heterocycl. Chem., 2016, 118, 47–90 CrossRef CAS.
- B. C. Das, M. Adil Shareef, S. Das, N. K. Nandwana, Y. Das, M. Saito and L. M. Weiss, Bioorg. Med. Chem., 2022, 63, 116748 CrossRef CAS PubMed.
- N. A. Schilling, A. Berscheid, J. Schumacher, J. S. Saur, M. C. Konnerth, S. N. Wirtz, J. M. Beltrán-Beleña, A. Zipperer, B. Krismer, A. Peschel, H. Kalbacher, H. Brötz-Oesterhelt, C. Steinem and S. Grond, Angew. Chem., Int. Ed., 2019, 58, 9234–9238 CrossRef CAS PubMed.
- S. Chatterjee and A. Bandyopadhyay, Org. Lett., 2023, 25, 2223–2227 CrossRef CAS PubMed.
- J. B. Mitra, S. Chatterjee, A. Kumar, A. Bandyopadhyay and A. Mukherjee, RSC Med. Chem., 2022, 13, 1239–1245 RSC.
- S. Chatterjee, A. Chowdhury, S. Saproo, N. Mani Tripathi, S. Naidu and A. Bandyopadhyay, Chem.–Eur. J., 2023, e202303327 Search PubMed.
- D. Li, Y. Chen and Z. Liu, Chem. Soc. Rev., 2015, 44, 8097–8123 RSC.
- G. Wulff, M. Lauer and H. Böhnke, Angew Chem. Int. Ed. Engl., 1984, 23, 741–742 CrossRef.
- N. Sahiba, A. Sethiya, J. Soni, D. K. Agarwal and S. Agarwal, Top. Curr. Chem., 2020, 378, 34 CrossRef CAS PubMed.
- S. Huh, N. Batistatou, J. Wang, G. J. Saunders, J. A. Kritzer and A. K. Yudin, RSC Chem. Biol., 2024, 5, 328–334 RSC.
- G. A. Ellis, M. J. Palte and R. T. Raines, J. Am. Chem. Soc., 2012, 134, 3631–3634 CrossRef CAS PubMed.
- S. Chatterjee, N. M. Tripathi and A. Bandyopadhyay, Chem. Commun., 2021, 57, 13629–13640 RSC.
- S. Sinha, P. Gaur, S. Dev, S. Mukhopadhyay, T. Mukherjee and S. Ghosh, Sens. Actuators, B, 2015, 221, 418–426 CrossRef CAS.
- S. Chatterjee, E. V. Anslyn and A. Bandyopadhyay, Chem. Sci., 2021, 12, 1585–1599 RSC.
- K. Li, W. Wang and J. Gao, Angew. Chem., Int. Ed., 2020, 59, 14246–14250 CrossRef CAS PubMed.
- H. Choi, M. Kim, J. Jang and S. Hong, Angew. Chem., Int. Ed., 2020, 59, 22514–22522 CrossRef CAS PubMed.
- F.-J. Chen and J. Gao, Chem.–Eur. J., 2022, 28, e202201843 CrossRef CAS PubMed.
- C. P. Ramil, P. An, Z. Yu and Q. Lin, J. Am. Chem. Soc., 2016, 138, 5499–5502 CrossRef CAS PubMed.
- A. Bandyopadhyay, S. Cambray and J. Gao, Chem. Sci., 2016, 7, 4589–4593 RSC.
- N. C. Rose, A. V. Sanchez, E. F. Tipple, J. M. Lynam and C. D. Spicer, Chem. Sci., 2022, 13, 12791–12798 RSC.
- P. G. Hains and P. J. Robinson, J. Proteome Res., 2017, 16, 3443–3447 CrossRef CAS PubMed.
- R. J. Spears, C. McMahon and V. Chudasama, Chem. Soc. Rev., 2021, 50, 11098–11155 RSC.
- F.-J. Chen, N. Pinnette, F. Yang and J. Gao, Angew. Chem., Int. Ed., 2023, e202306813 Search PubMed.
- A. Chowdhury, S. Chatterjee, A. Kushwaha, S. Nanda, T. J. Dhilip Kumar and A. Bandyopadhyay, Chem.–Eur. J., 2023, e202300393 CrossRef CAS PubMed.
- B. Halliwell, M. V. Clement and L. H. Long, FEBS Lett., 2000, 486, 10–13 CrossRef CAS PubMed.
- A. Bandyopadhyay, S. Cambray and J. Gao, J. Am. Chem. Soc., 2017, 139, 871–878 CrossRef CAS PubMed.
- H. M. Ellerby, W. Arap, L. M. Ellerby, R. Kain, R. Andrusiak, G. Del Rio, S. Krajewski, C. R. Lombardo, R. Rao, E. Ruoslahti, D. E. Bredesen and R. Pasqualini, Nat. Med., 1999, 5, 1032–1038 CrossRef CAS PubMed.
- L. Agemy, D. Friedmann-Morvinski, V. R. Kotamraju, L. Roth, K. N. Sugahara, O. M. Girard, R. F. Mattrey, I. M. Verma and E. Ruoslahti, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 17450–17455 CrossRef CAS PubMed.
- A. Tossi, L. Sandri and A. Giangaspero, Pept. Sci., 2000, 55, 4–30 CrossRef CAS.
- S. Hyun, S. Lee, S. Kim, S. Jang, J. Yu and Y. Lee, Biomacromolecules, 2014, 15, 3746–3752 CrossRef CAS PubMed.
- J. Li, K. Hu, H. Chen, Y. Wu, L. Chen, F. Yin, Y. Tian and Z. Li, Chem. Commun., 2017, 53, 10452–10455 RSC.
- A. Bandyopadhyay and J. Gao, J. Am. Chem. Soc., 2016, 138, 2098–2101 CrossRef CAS PubMed.
- T. Utsugi, A. J. Schroit, J. Connor, C. D. Bucana and I. J. Fidler, Cancer Res., 1991, 51, 3062–3066 CAS.
- I. D. Alves, M. Carré, M.-P. Montero, S. Castano, S. Lecomte, R. Marquant, P. Lecorché, F. Burlina, C. Schatz, S. Sagan, G. Chassaing, D. Braguer and S. Lavielle, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 2087–2098 CrossRef CAS PubMed.
- J. Connor, C. Bucana, I. J. Fidler and A. J. Schroit, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 3184–3188 CrossRef CAS PubMed.
- J. Lee, E.-T. Oh, H. Lee, J. Kim, H. G. Kim, H. J. Park and C. Kim, Bioconjugate Chem., 2020, 31, 43–50 CrossRef CAS PubMed.
- J. Lee, E.-T. Oh, Y. Joo, H. G. Kim, H. J. Park and C. Kim, New J. Chem., 2020, 44, 19734–19741 RSC.
- R. B. Mokhtari, T. S. Homayouni, N. Baluch, E. Morgatskaya, S. Kumar, B. Das and H. Yeger, Oncotarget, 2017, 8, 38022–38043 CrossRef PubMed.
- M. Michael, R. Garcia-Carbonero, M. M. Weber, C. Lombard-Bohas, C. Toumpanakis and R. J. Hicks, Oncologist, 2017, 22, 272–285 CrossRef CAS PubMed.
- W. H. Chen, X. D. Xu, G. F. Luo, H. Z. Jia, Q. Lei, S. X. Cheng, R. X. Zhuo and X. Z. Zhang, Sci. Rep., 2013, 3, 1–7 Search PubMed.
- B. K. Wagner, T. Kitami, T. J. Gilbert, D. Peck, A. Ramanathan, S. L. Schreiber, T. R. Golub and V. K. Mootha, Nat. Biotechnol., 2008, 26, 343–351 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc04348k |
| ‡ BKD and AC contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |