Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorinated trehalose analogues for cell surface engineering and imaging of Mycobacterium tuberculosis†
Collette S. Guy a,
James A. Gott
a,
James A. Gott b,
Jonathan Ramírez-Cárdenasc,
Christopher de Wolfa,
Christopher M. Furzea,
Geoff Westb,
Juan C. Muñoz-García
b,
Jonathan Ramírez-Cárdenasc,
Christopher de Wolfa,
Christopher M. Furzea,
Geoff Westb,
Juan C. Muñoz-García c,
Jesus Angulo
c,
Jesus Angulo c and
Elizabeth Fullam
c and
Elizabeth Fullam *a
*a
aSchool of Life Sciences, University of Warwick, Coventry, CV4 7AL, UK. E-mail: e.fullam@warwick.ac.uk; Tel: +44 (0)2476 574239
bWMG, University of Warwick, Coventry, CV4 7AL, UK
cInstituto de Investigaciones Químicas (IIQ), Consejo Superior de Investigaciones Científicas, Universidad de Sevilla, Avenida Américo Vespucio, 49, Sevilla, 41092, Spain
First published on 12th August 2024
Abstract
The sensitive, rapid and accurate diagnosis of Mycobacterium tuberculosis (Mtb) infection is a central challenge in controlling the global tuberculosis (TB) pandemic. Yet the detection of mycobacteria is often made difficult by the low sensitivity of current diagnostic tools, with over 3.6 million TB cases missed each year. To overcome these limitations there is an urgent need for next-generation TB diagnostic technologies. Here we report the use of a discrete panel of native 19F-trehalose (F-Tre) analogues to label and directly visualise Mtb by exploiting the uptake of fluorine-modified trehalose analogues via the mycobacterial trehalose LpqY-SugABC ATP-binding cassette (ABC) importer. We discovered the extent of modified F-Tre uptake correlates with LpqY substrate recognition and characterisation of the interacting sites by saturation transfer difference NMR coupled with molecular dynamics provides a unique glimpse into the molecular basis of fluorine-modified trehalose import in Mtb. Lipid profiling demonstrated that F-Tre analogues modified at positions 2, 3 and 6 are incorporated into mycobacterial cell-surface trehalose-containing glycolipids. This rapid one-step labelling approach facilitates the direct visualisation of F-Tre-labelled Mtb by Focused Ion Beam (FIB) Secondary Ion Mass Spectrometry (SIMS), enabling detection of the Mtb pathogen. Collectively, our findings highlight that F-Tre analogues have potential as tools to probe and unravel Mtb biology and can be exploited to detect and image TB.
Introduction
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), continues to threaten humanity and remains a major health challenge, having claimed over 1.3 million lives in 2022.1 Efforts to control the global TB pandemic have been jeopardised by the emergence of drug-resistant Mtb strains that are more complicated to treat and, in some instances, incurable.1,2 Early diagnosis is crucial for the global management of TB since the timely intervention with antitubercular treatment regimens has the potential to prevent transmission, limit the emergence and spread of antibiotic resistant strains and save lives, yet each year over 3.6 million individuals do not receive treatment compounding the TB burden.1,3,4 The current World Health Organisation approved TB diagnostics are limited to three main methods: microscopy, culture and nucleic acid amplification;3,5 however, the low-cost microscopy-based tests used most widely in low-resource settings have limitations, which include low sensitivity.6 Clearly, new molecular probes for improved, rapid, sensitive and accurate TB diagnostic technologies are needed.As the architecture of the Mtb cell envelope is distinct from other species, comprising an interconnected peptidoglycan-arabinogalactan-mycolic acid (mAGP) core interspersed with a diverse array of mycobacterial specific ‘free’ lipids and glycolipids,7–9 it provides a unique opportunity to develop Mtb pathogen specific reporter probes through utilisation of the specialised mycobacterial cell-wall assembly machinery for the direct incorporation of a detection moiety into the mycobacterial cell-envelope. Indeed, an array of fluorescent-based probes have been developed to monitor Mtb metabolism and image cell-envelope components, which have provided new insights into fundamental aspects of Mtb biology, including enzyme function, metabolism, cell wall biosynthesis and cell division.10–15 Since the Mtb cell envelope is highly abundant in trehalose-containing glycolipids that include the mycolic acid esters: trehalose monomycolates (TMM) and trehalose dimycolates (TDM), the acyltrehaloses comprising di- and polyacyltrehaloses and sulfoglycolipids, not typically found in other bacterial species,7–9 recent attention has focused on the use of labelled trehalose derivatives for the visualisation of Mtb.10–12,15–17 Incorporation of trehalose into mycobacterial cell-envelope glycolipids can proceed via two interlinked pathways: either through extracellular synthesis catalysed by the antigen 85 complex (Ag85)18 and/or trehalose import19 followed by biosynthesis of the TMM glycolipid within the cytoplasm (Fig. 1). The Ag85 complex, which include the secreted proteins Ag85A, Ag85B and Ag85C, is responsible for both the conjugation of extracellular mycolic acids to the arabinogalactan (AG) cell-wall core and the incorporation of a mycolic acid from one TMM into another TMM molecule to generate TDM.18 Alternatively the ‘free’ trehalose moiety, released concomitantly during Ag85 mediated cell-wall biosynthesis, can be recycled via the LpqY-SugABC ATP-binding cassette transporter19 and used as a precursor to yield cytoplasmic TMM, which is subsequently shuttled across the membrane by the MmpL3 exporter for insertion into the mycobacterial cell-envelope.20,21 Recent studies identified that both pathways can be hijacked to incorporate modified trehalose analogues into mycobacterial cell-envelope trehalose mycolates. The Ag85 complex is known to be particularly promiscuous for a wide panel of extensively modified trehalose substrates12 whereas the LpqY-SugABC importer only tolerates minor modifications.11,16,22 Recent structural and functional analysis of the LpqY substrate binding lipoprotein has afforded molecular level insights into the rules governing the precise substrate selectivity for this transporter enabling the design of reporter mimics to hijack this pathway.22
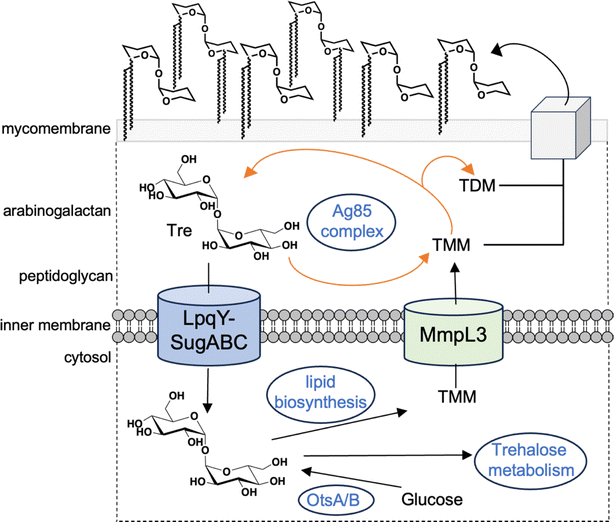 | ||
| Fig. 1 Schematic illustrating the pathways involved in trehalose processing in mycobacteria. TMM: trehalose monomycolate; TDM: trehalose dimycolate. | ||
To date, trehalose probes possessing ‘clickable’ azide and alkyne reactive handles have been used extensively for imaging of mycobacteria,11,12,16,23,24 however this approach requires extensive wash steps and a secondary, bulky fluorophore label for the direct visualisation of the cells, limiting their use. An 18F trehalose-modified radioprobe can be used to label the non-pathogenic model Mycobacterium smegmatis organism.17,25 However, this direct approach is challenging as fluorine-18 has a short half-life (∼109 minutes) and it is not possible to infer labelling of pathogenic Mtb because of the significant differences in nutrient uptake and processing between these diverse mycobacterial species.26–28 To address these potential limitations we rationalised we could leverage the Mtb LpqY-SugABC transporter to develop and deliver native 19F-modified trehalose reporter probes as a direct route to specifically visualise mycobacteria by focused ion beam (FIB) secondary ion mass spectrometry (SIMS), a technique which is increasingly being applied as a powerful tool to map elemental ions and molecules in biological samples.29–31 SIMs is an analytical technique that uses a focused ion beam to scan the sample surface, generating secondary ions detected by a mass analyser.32 Analysis of the mass spectrum generated on a pixel-by-pixel basis produces an ‘ion image’ of the composition, distribution and intensity of elements/molecules with spatial resolution within a sample.
We show here that a discrete panel of unnatural fluorine modified trehalose analogues are indeed tolerated by the Mtb LpqY-SugABC transporter and reveal the molecular architecture dictating their specific recognition. Subsequent characterisation demonstrated that this transport promiscuity can be harnessed to facilitate the uptake of fluorinated trehalose probes into live Mtb cells, which are subsequently incorporated into extracellular mycobacterial trehalose containing mycolic acids, enabling detection of the Mtb pathogen. Taken together, these results highlight the possibility of exploiting native fluorinated-trehalose derivatives to be used not only as tools for unravelling Mtb biology but also as new molecular probes for the direct visualisation and detection of Mtb.
Results and discussion
Panel of fluorinated trehalose analogues
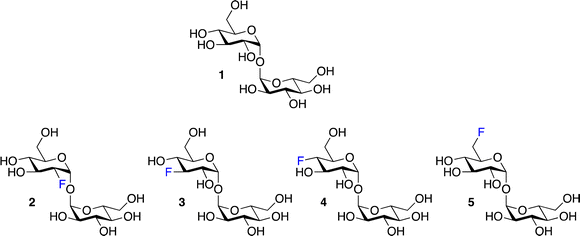
To explore the potential of fluorinated trehalose for Mtb detection and evaluate how selected modifications influence assimilation and cell-labelling, we synthesised a panel of fluorinated trehalose analogues with systematic modifications at either the 2-, 3-, 4-, or 6-position on one glucose subunit with retention of the natural stereochemistry (2–5). An established chemoenzymatic route33 was employed to access 2-deoxy-2-fluoro-trehalose (2F-Tre) (2), 3-deoxy-3-fluoro-trehalose (3F-Tre) (3) and 6-deoxy-6-fluoro-trehalose (6F-Tre) (5) in 42–49% yield, and a modified five-step synthetic route17,34 (Scheme S1†) afforded 4-deoxy-4-fluoro-trehalose (4F-Tre) (4), giving the focused panel shown in Fig. 2 (details provided in ESI, Schemes S1, Fig. S12–S30 and characterisation data†).LpqY substrate specificity for F-Tre analogues
As the mycobacterial specific LpqY-SugABC transporter is solely responsible for the import of trehalose,11,19,22 our first goal was to establish if the introduction, and position, of a fluorine moiety impacts on substrate recognition. We assessed the binding affinity of each fluorinated analogue (2–5) to LpqY, the substrate-binding domain that dictates substrate specificity and hence, subsequent transport, by microscale thermophoresis (MST). Highest binding affinities (Kd) were observed for 3F-Tre (110.8 ± 7.1 μM) and 6F-Tre (90.3 ± 0.9 μM) with comparable affinities to unmodified trehalose (109.5 ± 3.2 μM) whereas 2F-Tre and 4F-Tre bind ∼8- and ∼16-fold more weakly, with 4F-Tre having affinity in the millimolar range (Table 1, Fig. S1†). Altogether, these findings indicate that the position of the fluorine modification is key to LpqY recognition with replacement at the 3- or 6-positions preferred. This is in direct contrast to previous LpqY-azide-trehalose binding analyses (Table 1)22 that identified preferential binding of azide-trehalose modified at the 4- > 6- ≥ 3- > 2-positions, albeit with ∼2–4 weaker affinity than the corresponding fluorinated versions, highlighting the importance of both the substituent type and position in transporter-substrate tolerance and recognition.| Substrate | Kd (μM) | Reference |
|---|---|---|
| a Mean ± standard error mean is from at least three independent experiments. The substrate structures are shown in Fig. 2. The binding affinity data is shown in Fig. S1. | ||
| Trehalose | 109.5 ± 3.2 | This study |
| 2-Deoxy-2-fluoro-trehalose (2F-Tre) | 770.4 ± 23.2 | This study |
| 2-Deoxy-2-azido-trehalose (2Az-Tre) | 1915.6 ± 7.7 | 22 |
| 3-Deoxy-3-fluoro-trehalose (3F-Tre) | 110.8 ± 7.1 | This study |
| 3-Deoxy-3-azido-trehalose (3Az-Tre) | 474.9 ± 17.2 | 22 |
| 4-Deoxy-4-fluoro-trehalose (4F-Tre) | 1483.7 ± 75.6 | This study |
| 4-Deoxy-4-azido-trehalose (4Az-Tre) | 246.6 ± 7.1 | 22 |
| 6-Deoxy-6-fluoro-trehalose (6F-Tre) | 90.3 ± 0.9 | This study |
| 6-Deoxy-6-azido-trehalose (6Az-Tre) | 442.7 ± 5.3 | 22 |
Molecular basis of F-Tre analogue recognition
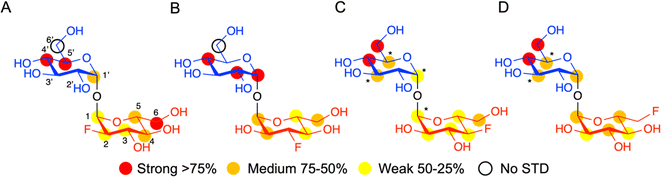
Having established that LpqY has the capacity to recognise fluorinated trehalose analogues we sought to determine the molecular rules of recognition in solution. Saturation transfer difference (STD) NMR confirmed binding for each fluorinated trehalose analogue, verifying that switching a hydroxyl group to fluorine at all possible positions is not detrimental for recognition by the mycobacterial trehalose transporter (Fig. 3). The STD NMR intensities differed depending on the temperature of the experiment (Fig. S2–S3 and Tables S1–S4†). More intense signal intensities were observed at low temperature (5 °C) for 2F-Tre and 4F-Tre, whereas a higher temperature of 30 °C was required for STD NMR observation of 3F-Tre and 6F-Tre, especially in the case of the 3-fluoro derivative where the particularly low signal-to-noise ratio meant no STD NMR analysis was possible at 5 °C. These results indicate a much stronger affinity for 3F-Tre and 6F-Tre, in comparison with the 2- and 4-fluoro derivatives, consistent with our binding affinity data (Table 1). STD binding epitope maps are unique for each F-Tre analogue (Fig. 3) and indicate that protons from both the unmodified and modified carbohydrate rings make distinct close contacts to LpqY relevant for recognition, with a significant overall decrease in the relative STD intensities at the modified fluorinated glucose ring (Fig. 3).Comparative DEEP-STD NMR analyses,35 which assess the type of protein residue that makes contacts with the fluorinated trehalose analogues (Fig. S4†), in combination with molecular dynamics simulations over 100 ns (Fig. S5†) revealed the relative orientations of the four ligands in the LpqY binding pocket (Fig. 4). Each of the F-Tre analogues bind in similar relative orientations and sit in an analogous position to trehalose (PDB 7APE)22 (Fig. S6†). The fluorinated-modified glucose ring is located towards the entrance of the substrate binding cavity, similar to the orientation we observed previously for 6Az-Tre,22 indicating that when the second glucose ring is modified it is preferentially accommodated within an expanded binding pocket at the channel entrance rather than buried at the base of the binding cleft (Fig. 4). Because the sidechains of Asn25, Glu26, Gln59 and Asn134 make hydrogen bond contacts with the modified glucose ring it is clear this interaction network plays an important role in controlling fluoro-modified trehalose specificity. Indeed, the significantly reduced affinity of 4F-Tre could be attributed to weakened interactions through the loss of a hydrogen-bond partner with Glu26 when a fluorine group is substituted at the 4-position.
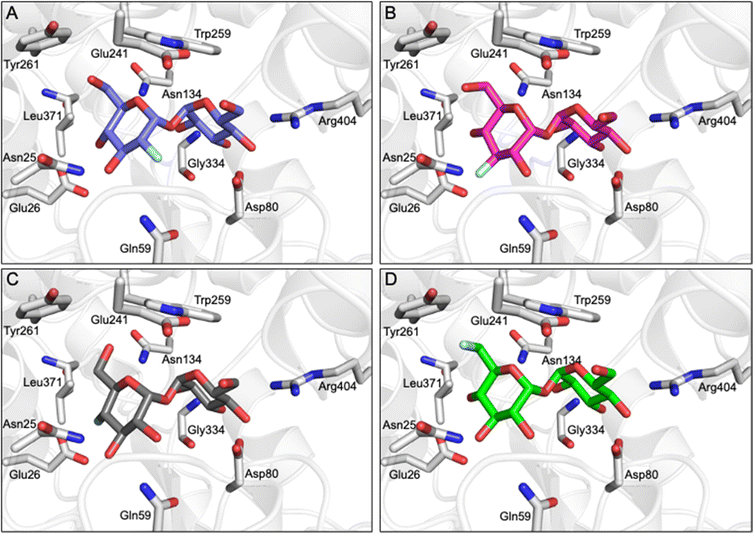 | ||
| Fig. 4 F-Tre binding to LpqY. Model of the mode of binding of F-Tre analogues based on experimentally derived DEEP-STD NMR derived interactions and molecular dynamic simulations. (A) 2F-Tre (blue carbon atoms), (B) 3F-Tre (magenta carbon atoms), (C) 4F-Tre (dark grey carbon atoms), and (D) 6F-Tre (green carbon atoms). Cartoon representation of LpqY in grey (PDB 7APE), trehalose and binding site residues are represented as sticks (oxygen: red; fluorine: pale blue). | ||
Mtb imports 2F-, 3F- and 6F-Tre analogues via the LpqY-SugABC transporter
Having confirmed that the fluorinated trehalose analogues bind to LpqY, and since not all substrates recognised by periplasmic binding domains are translocated by the cognate ABC transporter, we sought to evaluate whether these derivatives are taken up by live Mtb cells by ion chromatography. Treatment of Mtb cells with each fluorinated analogue led to a significant dose-dependent increase in the accumulation of 2F-Tre, 3F-Tre and 6F-Tre in Mtb cells, with the assimilation of micromolar levels even after exposure to the lowest concentration of the probe (25 μM) (Fig. 5). No uptake of 4F-Tre was detected, a finding consistent with its poor binding affinity to LpqY. The uptake pattern matches the entry of these analogues into non-pathogenic M. smegmatis cells, suggesting overlapping fluorine-modified trehalose analogue recognition profiles between these bacilli.17 These findings, together with the determined F-Tre analogue affinities for LpqY, emphasise the importance of the LpqY substrate binding domain in controlling which substrates are translocated.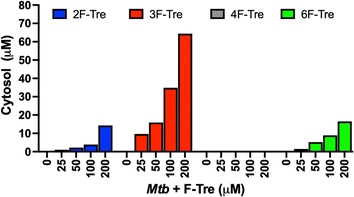 | ||
| Fig. 5 Ion chromatography analysis of F-Tre labelled Mycobacterium tuberculosis. Mtb was cultured in the presence of F-Tre analogues 2–5 (0–200 μM) and the cytosolic extracts analysed by high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD), and F-Tre concentration quantified. The ion-chromatography traces and retention times are shown in Fig. S7 and Table S5.† | ||
As Mtb efficiently imports the 2-, 3- and 6-fluorinated trehalose derivatives we then asked how rapidly each analogue is accumulated. We evaluated uptake in cytosolic extracts and calculated accumulation rates of 0.80 μM h−1 (2F-Tre) 0.94 μM h−1 (3F-Tre) and 0.82 μM h−1 (6F-Tre). Rapid uptake of each F-Tre derivative was observed at 30 min and at 8 hours ∼20% increased accumulation of 3F-Tre was detected compared to 2F- and 6F-Tre (Fig. 6, S8 and Table S6†). In agreement with previous studies that used radiolabelled 14C-trehalose,19 no saturation of uptake was observed for each F-Tre analogue 2, 3 and 5, indicating that an equilibrium between trehalose import and export has not yet been reached at the 8 hour timepoint, which may be consistent with the extremely slow growth rate of the Mtb bacillus or metabolic utilisation of the imported F-Tre analogues. Altogether, these observations establish that 2F-, 3F- and 6F-trehalose are rapidly transported, and accumulate, in live Mtb cells.
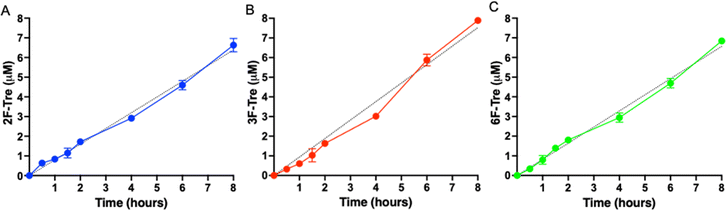 | ||
| Fig. 6 Time dependent uptake of F-Tre analogues in Mycobacterium tuberculosis. Mtb was cultured in the presence of (A) 2F-Tre, (B) 3F-Tre and (C) 6F-Tre (100 μM) and the cytosolic extracts sampled at the time points indicated and analysed by high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD). Error bars denote the standard deviation from duplicate experiments. Simple linear regression fit is shown with a black dotted line (A) y = 80x; (B) y = 0.94x; (C) y = 0.82x. The concentration values are in Table S6.† Representative ion-chromatography traces are shown in Fig. S8† and retention times are in Table S7.† | ||
We then asked whether the F-Tre analogues are imported via LpqY-SugABC transporter or via an alternative route and evaluated uptake in a Mycobacterium bovis BCG strain lacking the trehalose transporter (ΔlpqY-sugABC),11 as a model for Mtb. In agreement with our Mtb data 2F-, 3F- and 6F-Tre accumulate in wild-type M. bovis BCG cytosolic extracts and not 4F-Tre (Fig. 7). Notably, uptake of each F-Tre derivative was completely abolished in the ΔlpqY-sugABC mutant and transport restored in the corresponding complemented strain. Taken together, as the mutant strain is unable to utilise F-Tre, these data clearly show that uptake of fluorinated trehalose analogues occurs solely via the trehalose LpqY-SugABC import system.
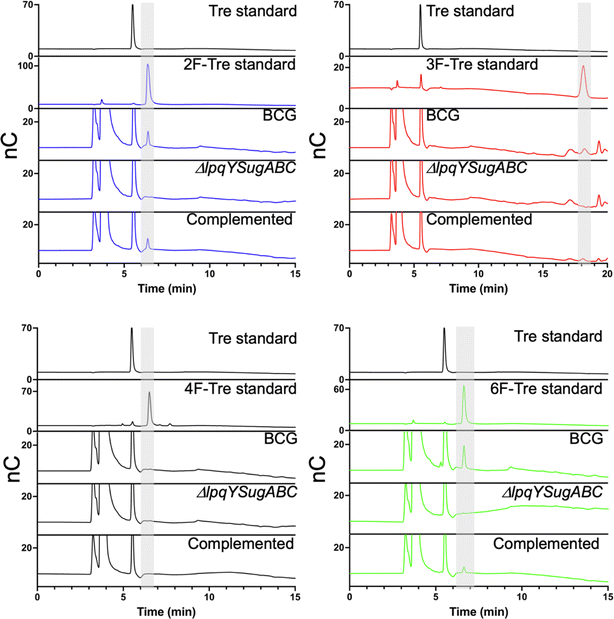 | ||
| Fig. 7 Comparison of F-Tre uptake in Mycobacterium bovis BCG and LpqY-SugABC mutant strains. Wild-type Mycobacterium bovis BCG and the ΔlpqY-sugABC Mycobacterium bovis BCG mutant and complemented strain Mycobacterium bovis BCG ΔlpqY-sugABC::lpqY-SugABC were cultured in the presence of F-Tre analogues 2, 3, 4 and 5 (100 μM) and the cytosolic extracts analysed by high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD). Retention times and peak areas are in Table S8.† | ||
Mtb incorporates 2F-, 3F- and 6F-Tre analogues into trehalose-containing glycolipids
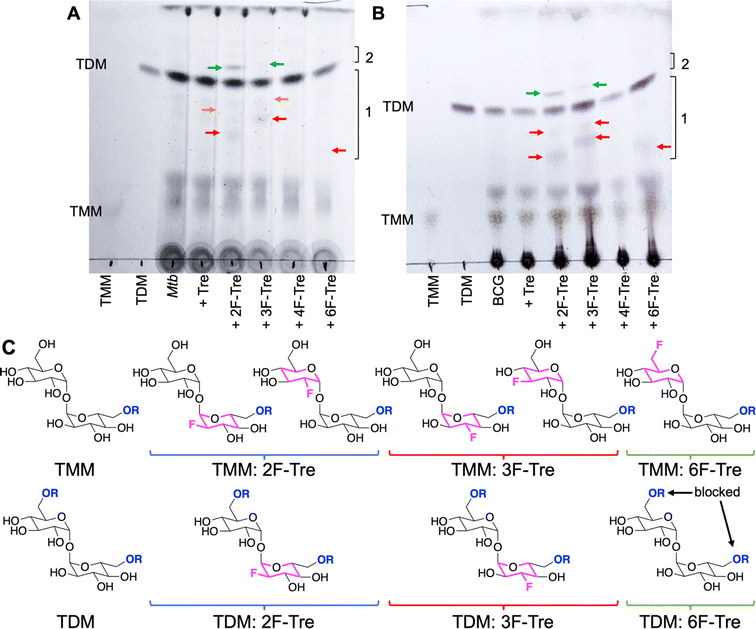
To determine the fate of F-Tre analogues following import across the mycobacterial cell-envelope Mtb and M. bovis BCG were exposed to each F-Tre analogue (2–5, 100 μM), alongside trehalose and untreated cells, and the extracted lipids evaluated by thin layer chromatography (TLC) and ion chromatography (Fig. 8, S9 and Table S9†). Treatment of Mtb and M. bovis BCG with 2F-, 3F- and 6F-Tre afforded new spots in the lipid extracts that were not present in the control samples (Fig. 8). No new spots were observed with 4F-Tre addition, consistent with its lack of uptake. Two additional lower polarity bands than TMM are present for the 2F- and 3F- samples and one for 6F-Tre (Fig. 8), indicating their incorporation into TMM (Fig. 8). This labelling pattern is consistent with two possible fluorinated-TMM regiochemistries for 2F- and 3F-Tre, where the fluorine is either on the same glucose moiety as the mycolic acid or the other subunit (Fig. 8). Since the TMM 6-position is blocked through mycolic attachment, only one 6 F trehalose isomer is possible, as observed (Fig. 8). The presence of one additional lower polarity TDM band for 2F- and 3F-Tre suggests these analogues are also incorporated into the TDMs. As expected, when mycobacteria are labelled with 6F-Tre no additional band in the TDM region is observed, since both 6-positions are inaccessible due to the attachment of mycolic acids on both glucose rings preventing the production of 6-fluorinated TDM.Altogether, these experiments demonstrate that, unless the 4-position is modified, mycobacteria have the capacity to take up fluorinated trehalose analogues and incorporate these modified sugars into cell-surface trehalose mycolates. It is notable that 4F-Tre is not found in any trehalose glycolipid in either Mtb or M. bovis BCG as this differs from the situation found for the analogous azido-trehalose derivatives which are all incorporated into mycobacterial glycolipids, albeit via different routes, depending on the mycobacterial species.11,17 For example, in M. bovis BCG, 3- and 4-azido trehalose are not recycled via LpqY-SugABC but, instead, incorporated into TMM and TDM by the extracellular Ag85 complex.11 Yet this does not occur for any F-Tre analogue. Thus, these findings reveal distinct mechanistic pathways for trehalose analogue recognition and use, which depend on both the type and position of modification and the mycobacterial species, highlighting an evolutionary divergence of trehalose transport across mycobacterial species.
Mtb does not readily metabolise 2F-, 3F- and 6F-Tre analogues into corresponding glucose analogues
Next, to determine if the fluorinated-trehalose derivatives are processed further we analysed the same cytosolic extracts of Mtb exposed to 2, 3, and 5 (100 μM) for the presence of fluorinated-glucose break-down products that would result from trehalase activity: 2-deoxy-2-fluoro-glucose (2F-Glc), 3-deoxy-3-fluoro-glucose (3F-Glc) and 6-deoxy-6-fluoro-glucose (6F-Glc) respectively (Fig. S10 and Table S10†). No cytosolic 3F- or 6F-glucose were found, indicating that the cleavage of 3F- and 6F-Tre does not occur. This suggests that intact 3F- and 6F-Tre are predominantly processed by pathways that ultimately incorporate these derivatives into cell-surface trehalose glycolipids. In contrast, Mtb labelled with 2F-Tre revealed the presence of low levels of 2F-Glc, indicating some degradation occurs, likely catalysed by the Mtb trehalase.36 It is possible that the 2F-Glc metabolite is then diverted into alternative glucose utilisation pathways and not solely incorporated into cell-surface trehalose opening up potential off-target labelling routes. As the F-Tre analogues effectively label Mtb glycolipids, we did not explore additional metabolic pathways. However the F-Tre analogues might also be metabolised by intracellular enzymes such as TreS, which isomerises trehalose to maltose, or through phosphorylation.Imaging F-Tre labelled Mtb by secondary ion mass spectrometry
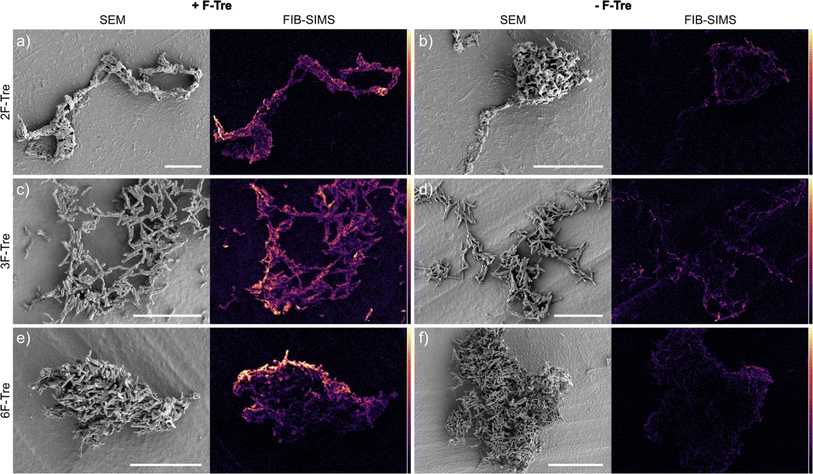
As fluorine-modified trehalose analogues are incorporated into cell-surface trehalose-containing Mtb glycolipids, we next examined the ability to visualise and image F-Tre labelled Mtb cells using focused ion beam (FIB) secondary ion mass spectrometry (SIMS), focused on 19F− ions. Elemental scanning for negative ions in the mass range 1–100 detected 19F− fluorine ions in all Mtb samples exposed to F-Tre and only trace levels in non-treated cells (Fig. S11† and 9). We then mapped the distribution of fluorine in the samples, first imaging the Mtb cells by scanning electron microscopy (SEM) to show the morphology of the cells and then analysis of the same section by FIB-SIMS. The intensity of the SIMS 19F− signal correlated with the scanning electron micrograph and showed a clear pattern of accumulation and localisation of F-Tre in treated Mtb cells (Fig. 9). In contrast only background 19F− levels were observed in untreated cells. Crucially, our findings establish that F-Tre labelling of live Mtb cells affords a prominent 19F− SIMS signal and low background demonstrating the feasibility of this approach for the direct detection and imaging of mycobacteria. Whilst robust labelling is observed for each probe, it is conceivable that the incorporation of multiple fluorine atoms on the same glucose ring that combines modifications at either the 2- 3-, or 6-positions could enhance the signal intensity further.Conclusions
One of the major hurdles in controlling the TB pandemic is the need for improved TB diagnosis. One route to achieve this is to transition from sputum microscopy to molecular testing. Yet molecular probes for the direct detection of TB are lacking. To address this deficiency, we used native fluorinated trehalose probes for imaging by FIB-SIMS, providing unique opportunities to specifically visualise and detect the Mtb pathogen. Given the requirement for specialised mass-spectrometry imaging equipment, optimisation will be necessary for use in resource limited settings. However 19-fluorine labelled probes may offer advantages over 18-fluorine methods, which have short half-lives, especially where point-of-care analysis is unfeasible and instead requires sample collection and central testing facilities. In conclusion, here we have clearly shown that we can hijack the promiscuity of the mycobacterial specific LpqY-SugABC trehalose transport machinery to incorporate fluorinated reporters into mycobacterial specific cell-surface glycolipids, combined with FIB-SIMS imaging to resolve 19F− localisation and directly image Mtb cells. Moreover, our findings demonstrate that F-Tre probes allow for the detection of the Mtb pathogen with high sensitivity. Expanding on this, our studies reveal a discernible dependence on the position of fluorine atom modification for efficient Mtb labelling. Specifically, we have uncovered the molecular framework that dictates probe recognition and import allowing us to explain F-Tre selectivity. We have shown that a single fluorine modification at the 2-, 3- and 6-positions of one glucose ring is optimal since substitution at the 4-position disrupts a critical hydrogen bond interaction that significantly weakens interactions with LpqY, impeding 4F-Tre import. The structural insights into F-Tre recognition and uptake analysis provided by this study, combined with FIB-SIMS imaging of Mtb cells provides a firm foundation that open opportunities for the rational development of high-contrast molecular probes for the detection of mycobacteria and provides an important step towards expanding the existing toolbox of molecular reporters for TB diagnosis.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Conceptulisation: CSG, EF; methodology: CSG, JAG, JRC, JCMG, CdW, EF; formal analysis: CSG, JAG, JRC, JCMG, JA, EF; investigation: CSG, JAG, JRC, GW, JCMG, CMF, EF; resources: JA; visualisation: CSG, JAG, JRC, JCMG, EF; supervision: GW, JCMG, JA, EF; writing - original draft: CSG , JAG, JRC, JCMG, JA, EF; writing - review & editing: CSG, JAG, JRC, CdW, CMF, GW, JCMG, JA, EF; project administration: JA, EF; funding acquisition: JA, EF.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a Sir Henry Dale Fellowship to EF jointly funded by the Wellcome Trust and Royal Society (104193/Z/14/Z and 104193/Z/14/B) and a research grant from the Leverhulme Trust (RPG-2019-087). We thank Dr Ben Swarts (Central Michigan University, USA) for providing the TreT construct, Professor Rainer Kalscheuer (HHU Dusseldorf, Germany) for kindly providing the M. bovis BCG mutant strains, Dr Saskia Bakker for help with sample preparation for FIB-SIMS and Dr James Harrison for critical reading of this manuscript. We acknowledge equipment access, training, and support made available by the Research Technology Facility (managed by Dr Sarah Bennett) of the Warwick Integrative Synthetic Biology Centre (WISB), which received funding from EPSRC and BBSRC (BB/M017982/1). JA and JCMG thank support from Ministerio de Ciencia e Innovación via the grant AEI/10.13039/501100011033/PID2022-142879NB-I00, co-funded by the European Regional Development Fund (ERDF) “A way of making Europe”.References
- WHO Global Tuberculosis Report, 2023, https://www.who.int/tb/publications/global_report/en/ Search PubMed.
- G. B. Migliori, S. Tiberi, A. Zumla, E. Petersen, J. M. Chakaya, C. Wejse, M. Munoz Torrico, R. Duarte, J. W. Alffenaar, H. S. Schaaf, B. J. Marais, D. M. Cirillo, R. Alagna, A. Rendon, E. Pontali, A. Piubello, J. Figueroa, G. Ferlazzo, A. Garcia-Basteiro, R. Centis, D. Visca, L. D'Ambrosio, G. Sotgiu and N. members of the Global Tuberculosis, MDR/XDR-TB management of patients and contacts: Challenges facing the new decade. The 2020 clinical update by the Global Tuberculosis Network, Int. J. Infect. Dis., 2020, 92, S15–S25 CrossRef CAS PubMed.
- M. Pai, P. K. Dewan and S. Swaminathan, Transforming tuberculosis diagnosis, Nat. Microbiol., 2023, 8, 756–759 CrossRef CAS PubMed.
- M. Pai and C. Raison, Transforming the diagnosis of tuberculosis: an editorial board member's opinion at the 15th year of Expert Review of Molecular Diagnostics, Expert Rev. Mol. Diagn., 2015, 15, 295–298 CrossRef PubMed.
- WHO Standard: Universal Access to rapid tuberculosis diagnostics, https://www.who.int/publications/i/item/9789240071315 Search PubMed.
- The unsustainable anachronism of tuberculosis diagnosis, Lancet Microbe, 2023, 4, e379 Search PubMed.
- K. A. Abrahams and G. S. Besra, Mycobacterial cell wall biosynthesis: a multifaceted antibiotic target, Parasitology, 2018, 145, 116–133 CrossRef PubMed.
- P. J. Brennan, Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis, Tuberculosis, 2003, 83, 91–97 CrossRef CAS PubMed.
- M. Jackson, The mycobacterial cell envelope-lipids, Cold Spring Harbor Perspect. Med., 2014, 4, a021105 CrossRef PubMed.
- H. L. Hodges, R. A. Brown, J. A. Crooks, D. B. Weibel and L. L. Kiessling, Imaging mycobacterial growth and division with a fluorogenic probe, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5271–5276 CrossRef CAS PubMed.
- B. M. Swarts, C. M. Holsclaw, J. C. Jewett, M. Alber, D. M. Fox, M. S. Siegrist, J. A. Leary, R. Kalscheuer and C. R. Bertozzi, Probing the mycobacterial trehalome with bioorthogonal chemistry, J. Am. Chem. Soc., 2012, 134, 16123–16126 CrossRef CAS PubMed.
- K. M. Backus, H. I. Boshoff, C. S. Barry, O. Boutureira, M. K. Patel, F. D'Hooge, S. S. Lee, L. E. Via, K. Tahlan, C. E. Barry III and B. G. Davis, Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis, Nat. Chem. Biol., 2011, 7, 228–235 CrossRef CAS PubMed.
- A. A. Pohane, C. R. Carr, J. Garhyan, B. M. Swarts and M. S. Siegrist, Trehalose Recycling Promotes Energy-Efficient Biosynthesis of the Mycobacterial Cell Envelope, mBio, 2021, 12(1) DOI:10.1128/mbio.02801-20.
- A. Garcia-Heredia, A. A. Pohane, E. S. Melzer, C. R. Carr, T. J. Fiolek, S. R. Rundell, H. C. Lim, J. C. Wagner, Y. S. Morita, B. M. Swarts and M. S. Siegrist, Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria, eLife, 2018, 7, e37243 CrossRef PubMed.
- M. Kamariza, P. Shieh, C. S. Ealand, J. S. Peters, B. Chu, F. P. Rodriguez-Rivera, M. R. Babu Sait, W. V. Treuren, N. Martinson, R. Kalscheuer, B. D. Kana and C. R. Bertozzi, Rapid detection of Mycobacterium tuberculosis in sputum with a solvatochromic trehalose probe, Sci. Transl. Med., 2018, 10, eaam6310 CrossRef PubMed.
- H. L. Parker, R. M. F. Tomas, C. M. Furze, C. S. Guy and E. Fullam, Asymmetric trehalose analogues to probe disaccharide processing pathways in mycobacteria, Org. Biomol. Chem., 2020, 18, 3607–3612 RSC.
- S. R. Rundell, Z. L. Wagar, L. M. Meints, C. D. Olson, M. K. O'Neill, B. F. Piligian, A. W. Poston, R. J. Hood, P. J. Woodruff and B. M. Swarts, Deoxyfluoro-d-trehalose (FDTre) analogues as potential PET probes for imaging mycobacterial infection, Org. Biomol. Chem., 2016, 14, 8598–8609 RSC.
- J. T. Belisle, V. D. Vissa, T. Sievert, K. Takayama, P. J. Brennan and G. S. Besra, Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis, Science, 1997, 276, 1420–1422 CrossRef CAS PubMed.
- R. Kalscheuer, B. Weinrick, U. Veeraraghavan, G. S. Besra and W. R. Jacobs Jr, Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 21761–21766 CrossRef CAS PubMed.
- C. Varela, D. Rittmann, A. Singh, K. Krumbach, K. Bhatt, L. Eggeling, G. S. Besra and A. Bhatt, MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria, Chem. Biol., 2012, 19, 498–506 CrossRef CAS PubMed.
- Z. Xu, V. A. Meshcheryakov, G. Poce and S. S. Chng, MmpL3 is the flippase for mycolic acids in mycobacteria, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 7993–7998 CrossRef CAS PubMed.
- C. M. Furze, I. Delso, E. Casal, C. S. Guy, C. Seddon, C. M. Brown, H. L. Parker, A. Radhakrishnan, R. Pacheco-Gomez, P. J. Stansfeld, J. Angulo, A. D. Cameron and E. Fullam, Structural basis of trehalose recognition by the mycobacterial LpqY-SugABC transporter, J. Biol. Chem., 2021, 296, 100307 CrossRef CAS PubMed.
- H. N. Foley, J. A. Stewart, H. W. Kavunja, S. R. Rundell and B. M. Swarts, Bioorthogonal Chemical Reporters for Selective In Situ Probing of Mycomembrane Components in Mycobacteria, Angew Chem. Int. Ed. Engl., 2016, 55, 2053–2057 CrossRef CAS PubMed.
- A. A. Pohane, D. J. Moore, I. Lepori, R. A. Gordon, T. O. Nathan, D. M. Gepford, H. W. Kavunja, B. M. Swarts and M. S. Siegrist, A Bifunctional Chemical Reporter for In Situ Analysis of Cell Envelope Glycan Recycling in Mycobacteria, ACS Infect. Dis., 2022, 8, 2223–2231 CrossRef CAS PubMed.
- S. Pena-Zalbidea, A. Y. Huang, H. W. Kavunja, B. Salinas, M. Desco, C. Drake, P. J. Woodruff, J. J. Vaquero and B. M. Swarts, Chemoenzymatic radiosynthesis of 2-deoxy-2-[(18)F]fluoro-d-trehalose ([(18)F]-2-FDTre): A PET radioprobe for in vivo tracing of trehalose metabolism, Carbohydr. Res., 2019, 472, 16–22 CrossRef CAS PubMed.
- M. Niederweis, Nutrient acquisition by mycobacteria, Microbiology, 2008, 154, 679–692 CrossRef CAS PubMed.
- M. Braibant, P. Gilot and J. Content, The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis, FEMS Microbiol. Rev., 2000, 24, 449–467 CrossRef CAS PubMed.
- D. J. Beste, K. Noh, S. Niedenfuhr, T. A. Mendum, N. D. Hawkins, J. L. Ward, M. H. Beale, W. Wiechert and J. McFadden, 13C-flux spectral analysis of host-pathogen metabolism reveals a mixed diet for intracellular Mycobacterium tuberculosis, Chem. Biol., 2013, 20, 1012–1021 CrossRef CAS PubMed.
- D. Debois, K. Hamze, V. Guerineau, J. P. Le Caer, I. B. Holland, P. Lopes, J. Ouazzani, S. J. Seror, A. Brunelle and O. Laprevote, In situ localisation and quantification of surfactins in a Bacillus subtilis swarming community by imaging mass spectrometry, Proteomics, 2008, 8, 3682–3691 CrossRef CAS PubMed.
- P. Agüi-Gonzalez, S. Jähne and N. T. N. Phan, SIMS imaging in neurobiology and cell biology, J. Anal. At. Spectrom., 2019, 34, 1355–1368 RSC.
- A. Fearns, D. J. Greenwood, A. Rodgers, H. Jiang and M. G. Gutierrez, Correlative light electron ion microscopy reveals in vivo localisation of bedaquiline in Mycobacterium tuberculosis-infected lungs, PLoS Biol., 2020, 18, e3000879 CrossRef CAS PubMed.
- G. L. Fisher, A. L. Bruinen, N. Ogrinc Potocnik, J. S. Hammond, S. R. Bryan, P. E. Larson and R. M. Heeren, A New Method and Mass Spectrometer Design for TOF-SIMS Parallel Imaging MS/MS, Anal. Chem., 2016, 88, 6433–6440 CrossRef CAS PubMed.
- B. L. Urbanek, D. C. Wing, K. S. Haislop, C. J. Hamel, R. Kalscheuer, P. J. Woodruff and B. M. Swarts, Chemoenzymatic synthesis of trehalose analogues: rapid access to chemical probes for investigating mycobacteria, ChemBioChem, 2014, 15, 2066–2070 CrossRef CAS PubMed.
- R. W. Bassily, R. I. el-Sokkary, B. A. Silwanis, A. S. Nematalla and M. A. Nashed, An improved synthesis of 4-azido-4-deoxy- and 4-amino-4-deoxy-alpha,alpha-trehalose and their epimers, Carbohydr. Res., 1993, 239, 197–207 CrossRef CAS PubMed.
- S. Monaco, L. E. Tailford, N. Juge and J. Angulo, Differential Epitope Mapping by STD NMR Spectroscopy To Reveal the Nature of Protein-Ligand Contacts, Angew Chem. Int. Ed. Engl., 2017, 56, 15289–15293 CrossRef CAS PubMed.
- J. D. Carroll, I. Pastuszak, V. K. Edavana, Y. T. Pan and A. D. Elbein, A novel trehalase from Mycobacterium smegmatis - purification, properties, requirements, FEBS J., 2007, 274, 1701–1714 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc00721b |
| This journal is © The Royal Society of Chemistry 2024 |