Open Access Article
Open Access ArticlePhenylboronic acid-derived nanovectors for gene/drug delivery by targeting cell surface glycans
Venkanna Muripiti *a,
Venkatesh Ravulab,
Srinivas Batthulac,
Janardhan Banothu
*a,
Venkatesh Ravulab,
Srinivas Batthulac,
Janardhan Banothu d and
Ramesh Gondru*e
d and
Ramesh Gondru*e
aDepartment of Education, Central University of Kerala, Tejaswini Hills, Kasaragod-671320, Kerala, India. E-mail: muripitivenkanna@cukerala.ac.in
bLaboratory of Self Assembled Biomaterials and Translational Science, Institute for Stem Cell Science and Regenerative Medicine (DBT-inStem), GKVK Post, Bellary Road, Bengaluru, 560065, Karnataka, India
cDepartment of Pharmacy, Kangwon National University, Hyoja2-dong, Chuncheon, Republic of Korea
dDepartment of Chemistry, National Institute of Technology Calicut, Kozhikode-673601, Kerala, India
eFood Chemistry Division, ICMR–National Institute of Nutrition (NIN), Hyderabad-500007, Telangana, India. E-mail: ramesh.gondru@icmr.gov.in
First published on 26th March 2024
Abstract
Gene mutations within cells can lead to cancer, a global health challenge affecting millions worldwide. In combating cancer, various treatments such as surgery, radiotherapy, and chemotherapy have been employed. However, the distinct underlying genetic abnormalities causing the cancer are sometimes not addressed by conventional treatments. Adding to these obstacles, targeted therapy is another continuing challenge in cancer treatment. According to recent reports, phenylboronic acid (PBA)-decorated nanoparticles efficiently transfer genes to the intended location due to their strong affinity for sialic acid (SA), which is typically overexpressed in cancerous cells. These PBA-decorated nanoparticles may connect to cancer cells specifically, which enables them to target and deliver the cargo to cancer cells. Therefore, the present review concentrates on the role of PBA-decorated nanoparticles in gene/drug delivery. It includes a discussion on various boronic acid (BA)-conjugated macromolecules. We begin with an exploration of the chemistry underlying BA and its utility in effective delivery. Furthermore, the review elaborates on its application as a targeting ligand, providing a promising avenue for more precise and effective cancer treatment strategies.
1. Introduction
Cancer is the collective name for a wide variety of diseases that are caused by the uncontrolled proliferation of malignant cells. All cancers begin with faulty proteins involved in the cell's regeneration process that are encoded by mutated genes (oncogenes).1 The World Health Organization (WHO) reported cancer as the second largest cause of death worldwide after cardiovascular diseases (CVDs), with an estimated 10 million deaths each year, a number estimated to reach ∼30 million in 2040.2–4 Currently, chemotherapy is used to cure cancer by killing or inhibiting the proliferation of cancerous cells. However, lack of specificity, the drug resistance of cancer cells, and significant side-effects from killing normal cells limit the therapeutic potency of most available chemotherapeutic drugs.5 Therefore, early detection of cancer cells and the development of new cancer diagnostic methods and novel anticancer agents (with maximum efficacy and low toxicity) are essential for early treatment and speedy recovery.6 Most of the current traditional methods rely on the identification of tumor-related biomarkers. The major part of these biomarkers include cell surface glycoproteins and their components such as cancer antigen 125 (CA125), cluster of differentiation 44 (CD44), prostate-specific membrane antigen (PSMA), and Mucin1 (MUC1).7–11Among the many sugars that make up glycoproteins on the cell surface, sialic acids (SAs) located at the terminal end of glycans are of critical importance in the detection and therapy of various cancers.12,13 SAs are nine-carbon backbone monosaccharides usually found as terminal residues of cell surface glycoconjugates (glycoproteins and glycolipids)14 and are closely associated with several diseases, including cancers/tumors.15,16 The distribution of SAs is ubiquitous, being secreted as glycoconjugates in vertebrates and ‘higher’ invertebrates, and they can mediate or modulate a variety of physiological17 and pathological processes (infection and survival of pathogens), and help with cell–cell and cell–pathogen communication, including cell adhesion and immune responses.18 SAs have many structural and modulatory roles due to their negative charge and hydrophilicity.19 They act as a component of binding sites for various pathogens and toxins, and finally, by ‘molecular mimicry’, allow successful pathogenic microbes to adhere with SAs thus abetting host immune evasion.14,20–25 For instance, hemagglutinin (HGN), a protein found on the surface of influenza viruses, binds to SAs in human respiratory cells. This binding initiates the infection process. However, different influenza strains can adhere to different types of SAs due to differences in their hyaluronic acid (HA) structures. It is worth noting that certain strains can change to develop additional SA-binding capabilities, potentially resulting in new epidemics.26
SA overexpression is a common manifestation in cancer-associated cells for their progression, poor prognosis, and high metastatic potential. Hence, monitoring, analyzing, and controlling cancer cells through the recognition of specific binding sites of SA would be essential in diagnostic and therapeutic applications.12,27–30 However, efficacy and selectivity in SA on the tumor cell surface remain a challenge to the scientific community.2 SA has a complicated and intricate involvement in cancer genesis, progression, and therapy. Cancer cells frequently have increased sialic acid expression in comparison with healthy cells. This increase may be attributed to certain enzymes involved in SA production (sialyltransferases).31 Sialic acids enhance adhesion and motility by allowing cells to interact with other molecules in the tumor microenvironment. This can promote tumor invasion and metastasis.32 Increased sialylation protects cancer cells from immune cell detection and attack. This enables tumors to evade immune monitoring and grow unrestrained. SAs can contribute to drug resistance by inhibiting the binding and absorption of therapeutic medicines at their target locations on cancer cells.31
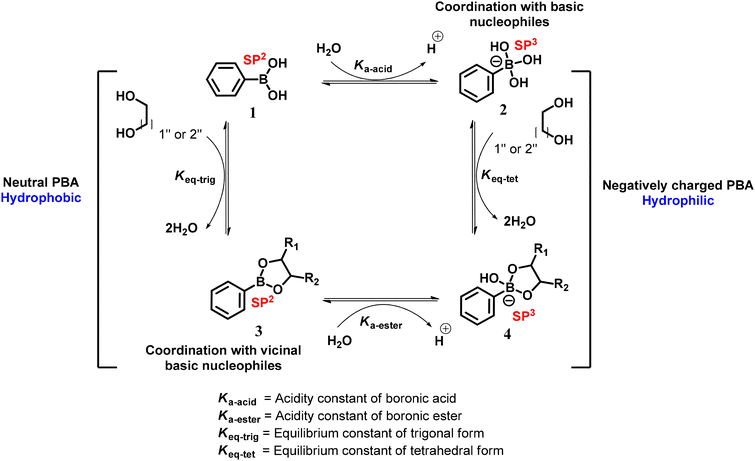
Presently, attention in research is directed towards non-traditional chemotherapies, such as targeted drug delivery, since they have the potential to boost the drug's bioavailability to tumor tissues.33 In recent years, targeted drug delivery systems (DDSs) have gained immense attention due to their enhanced biocompatibility with cancer cells,34,35 rich specific surface areas, and reduced metastases.36 Among them, boronic acid (BA) and its derivatives have attracted considerable attention as target substances due to their specificity and selectivity towards SAs on the tumor surface.37,38 In addition, at pH 8.8, BA forms a cyclic trigonal planar structure with SA, which can be made reversible at lower pH. This attribute can be exploited for drug delivery39,40 (Fig. 1). Lewis bases contribute electrons to boric acids because of their empty orbitals. SA acts as a Lewis base because its hydroxyl groups deprotonate at around pH 8.8, acquiring a negative charge and increased electron-donating capacity. The interaction of SAs’ negatively charged hydroxyl groups with the empty orbital of boron generates a reversible covalent bond. The geometry of the coordinating atoms and boron usually results in a cyclic trigonal planar structure. The fact that this interaction is pH-dependent is critical in drug delivery. The deprotonated hydroxyl groups cause significant interaction between BA and SA at physiological pH (about 7.4). This permits the boron-based chemical to transport a drug molecule linked to it. However, when the complex enters the cell's slightly more acidic environment (about pH 6.8), protonation of the hydroxyl groups weakens the BA–SA connection. This causes the drug molecule to be released into the mammalian cell's cytoplasm.40,41
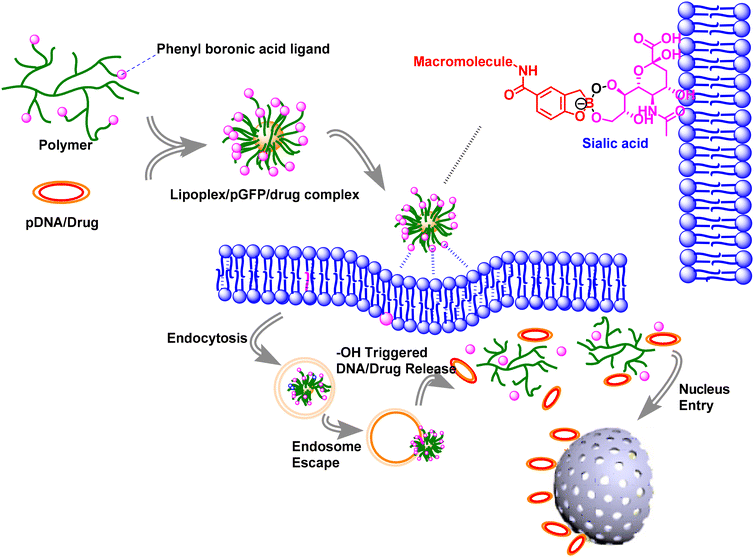 | ||
| Fig. 1 The schematic paradigm of the gene/drug delivery system showing crucial aspects of understanding the delivery process. | ||
BA is less toxic, acts as a Lewis acid and it is biocompatible.42–44 It has been known that sialylated epitopes on the surface of cancer cells can selectively interact with BA and its derivatives. This interaction can effectively control cancer cell adherence and recognition.45,46 Additionally, BA-based nanoparticles exhibit greater stability throughout circulation in the bloodstream and adequate uptake by target cells.47,48 This review offers a comprehensive insight into the latest applications of SA-targeted biomaterials, encompassing (1) the significance of BA chemistry in drug/gene delivery, (2) the functionality of phenylboronic acid (PBA) nanoparticles employed in drug/gene delivery, (3) hybrid vectors combining BA and peptides for gene/drug delivery, and (4) hybrid nanoparticles comprising polymer-conjugated PBA vectors for the delivery of siRNA, drugs, and genes. Finally, we outline a framework for the future progress in SA-targeted biomaterials.
2. Role of boronic acid (BA) chemistry in drug/gene delivery
Boronic acids (BAs) are commonly referred to as Lewis acids, with pKa values of 4–10.38,49 The pKa values vary depending on the type of substituents on the aromatic ring of BA as follows: if an electron-withdrawing group (EWG) is present, the pKa value will decrease, while if an electron-donating group (EDG) is present, the pKa value will increase.49 BA and its derivatives bind covalently to cis-1,2 or 1,3-diols (common motifs of sugars) with high affinity through reversible five- or six-membered boronate ester formation in an aqueous medium (Fig. 2). Because of the sugar-binding ability of BAs, there have been studies on synthetic mimics of carbohydrate-binding proteins (lectins), termed ‘borono-lectins’.40,50–53 Also, such binding fosters use of BAs as recognition moieties in the development of carbohydrate, fluorometric, and calorimetric sensors and artificial receptors.37,54BA and its ester usually exist in a neutral trigonal planar form (at physiological pH) similar to the carbocation.42,55 Also, the central boron atom is isoelectronic with the carbon cation and both have an empty p-orbital. In contrast to a carbocation, the mild Lewis acidic nature of the boron atom readily allows establishing reversible covalent bonds with nucleophiles like oxygen found in reactive oxygen species (ROS) and nitrogen in aqueous environments at a pH value higher than the pKa.56 This Lewis acid–base reaction results in the interconversion of the central boron atom from an uncharged trigonal planar57 (trig) geometry to an anionic tetrahedral (tet) structure, which is iso-electronically equivalent to a neutral sp3-hybridized quaternary carbon.58,59 Hence, the equilibrium between the two forms (neutral-sp2 and ionized-sp3) depends on the pKa values of BAs. The tumor microenvironment acts as a reduced buffer, donating electrons, while boron (in BA) itself acts as a Lewis acid; hence BA-containing nanoparticles act as Lewis acids, causing a shift in the boronic acid structure to the anionic tetrahedral type.60,61 BA-based nanoparticles with a specific morphology influence the release of the drug/gene from the nanoparticle into the cytosol62,63 (Fig. 1).
3. Functions of phenylboronic acid (PBA) nanoparticles in drug/gene delivery
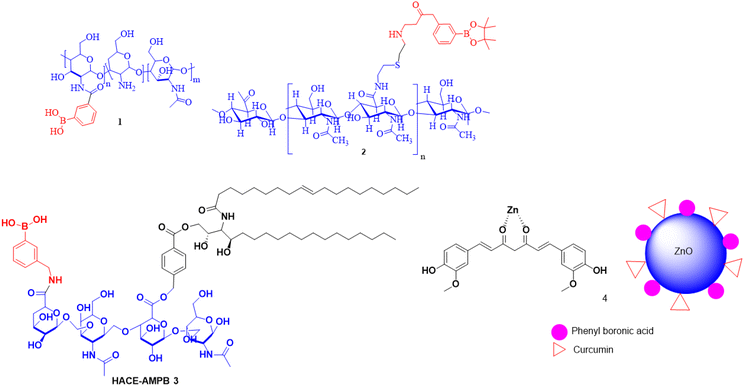
Over the past few decades, nanotechnology has been extensively studied and exploited increasingly for use in cancer therapy, including applications in diagnosis and precise targeting. It can play a significant role in DDSs and has the potential to overcome various drawbacks associated with conventional formulations and hence can revolutionize the approaches to drug delivery for meticulous therapeutic procedures.64–66 PBA and its derivatives as target substances are the most studied as they specifically recognize SAs that are aberrantly expressed (hyper sialylation) on the tumor cell surface during tumor transformation and malignant progression.27,67 In addition, less-toxic and non-immunogenic properties of PBA make it an attractive ligand for targeting tumor cells. Compared to conventional drugs, the interaction of surface-functionalized NPs with SA has the potential to achieve precise targeting of malignant tissues/cells, by enhancing permeability, transport across biological barriers, biocompatibility and stability.68,69Qiao Tang et al. reported PBA-derived lipid NPs for messenger RNA (mRNA) delivery and CRISPR/Cas9 genome editing. The experimental results revealed that PBA–BADP/mRNA NPs showed 300 times higher luciferase activity for reporter-gene expression in cancer cells compared to noncancerous cells. Furthermore, they demonstrated that PBA–BADP (Fig. 3) selectively prohibits cancer cell growth when it delivers the p53 mRNA gene, while delivery of PBA–BADP/Cas9 mRNA NPs knocks out the gene-expression of Henrietta Lacks (HeLa) cells with maximum efficacy than in normal cells.70 Ruonan Wang et al. reported an interesting finding that PBA modification can greatly expedite the lysosomal escape of cylindrical polymer brushes (CPBs) and further promote their exocytosis and transcellular translocation. They hypothesized that the mechanism of PBA-augmented lysosomal escape is linked to specific interactions of the PBA group with the lysosomal membrane proteins and heat shock proteins (HSPs). They also stated that their PBA modification has a prominent edge over the known lysosomal escape strategies in that the former does not cause notable detrimental effects on the properties of the CPBs (Fig. 3); on the contrary, it enhances their tumor accumulation and penetration exponentially.71
Won Jung Kim et al. developed a glucose-responsive continuous insulin (GRCI) release system by creating a novel design of polyhedral oligosilsesquioxane (POSS) modified with 3-aminophenylboronic acid (APBA). This modification not only resulted in a high insulin entrapment efficiency of 73.2% (Fig. 3), but also ensured a rapid response, effective distribution, and outstanding biocompatibility. Furthermore, they explored the effective release of insulin stimulated by glucose from the POSS–APBA micelles at neutral pH.72
4. Boronic acid–peptide hybrid vectors for gene/drug delivery
Peptides have advantageous properties by nature, including biocompatibility, adequate water solubility, and a relatively simple method of production (solid-phase peptide synthesis, for example).73 Peptide-based vectors are generally less toxic and mimic the structure of protein α-helices and β-sheets.74 Hence, peptides transport nucleic acids into cells by overcoming biological barriers such as cellular absorption, endosomal escape, and nuclear threshold.75 In order to find peptide ligands with unique biological activities, boronic acid has thus been extensively combined with peptides, an endeavor that has produced a wide range of applications. Using the combination of boronic acid and peptides as a starting point, many research groups76 have given a summary of recent developments in the field and have emphasized the adaptable and strong properties of boronic acid. Due to their ability to bond covalently with sugars or amino acids, peptide boronic acids are commonly used as ligands that are carriers of RNA and glycans, and covalently changeable inhibitors of enzymes.77–79 Xue-Long Sun's research group developed a boronic acid–lectin-based vector that formed a reversible boronate ester when it combined with 1,2- and 1,3-diols of saccharides in aqueous environments.80 Their biological investigation demonstrated that through inhibiting the lectin–FITC ensemble, the BSA–PBA combination attaches to the SA on the cell outside of Raw 264.7 cells.80 cRGD peptides and a polyethyleneimine–polyethylene glycol polymer are used to adorn the surface of boronic acid-rich bovine serum albumin nanoparticles, which have sizes of 70 nm, 110 nm, and 150 nm. Researchers have shown that incorporating a boronic acid group into bovine serum albumin nanoparticles increases particle size and enhances the surface of the particles, which can significantly boost both the concentration of drugs delivered and the duration of particle presence at the tumor site.60 Webster L. Santos's research group has identified branched peptide boronic acids (BPBAs) that bind to RRE IIB.81 In both eukaryotic cell lines, BPBA1 demonstrated good binding affinity, reasonable cell permeability, and low cytotoxicity. Nevertheless, it was unable to prevent the development of HIV-1 p24 capsids.81 However, it was eventually shown shortly after that BPBA3 effectively and dose-dependently inhibited the synthesis of HIV-1 p24 capsids, with an IC50 value of approximately 5 μM. According to theory, BPBA3 may cause the tertiary structure of the internal loop to alter conformation, exposing the RNA to RNase cleavage and inhibiting the replication of HIV-1.82 Yoshihiro Furukawa led a team for the development of boron-assisted abiotic polypeptide synthesis. Their experimental results revealed that under these circumstances, initial proteins and RNAs could interact and produce RNAs, which could then be passed down through RNA-dependent protein production to subsequent generations.83 In order to create a tumor-targeted gene carrier, PBA-functionalized PAMAM (PPP), for Bcl-2 siRNA delivery, phenylboronic acid (PBA) and amine-terminated polyamidoamine (PAMAM) were successfully conjugated in a study utilizing the heterobifunctional crosslinker NHS-PEG5k-Mal.845. Hybrid nanoparticles of polymer-conjugated phenylboronic acid vectors for the delivery of siRNA, drugs, and genes
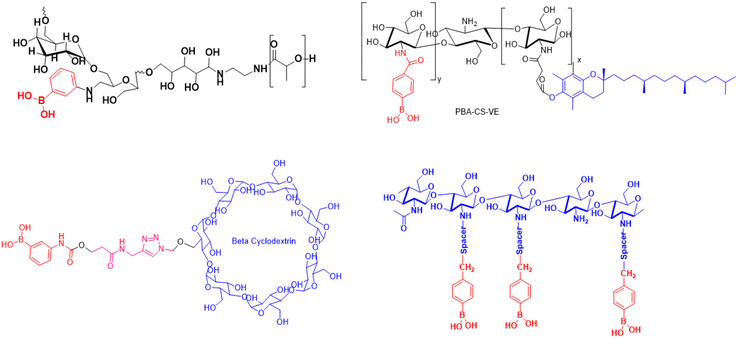
PBA can interact with guanidinium groups found on proteins, forming cationic connections with their assistance.85 Scientists from East China Normal University in China have discovered PBA-rich dendrimers for cytosolic transport using various model natural proteins, including cas9 ribonucleoprotein86 (Fig. 4). P4 outperforms dendrimers in efficiency tests, while P7, similar to P4, performs poorly in delivering BSA–FITC due to decreased dendrimer attachment effectiveness. Due to intramolecular nitrogen–boronate coordination, P8 demonstrated high translocation efficiency in BSA–FITC delivery, highlighting the crucial role of phenyl groups in protein binding and cytosolic delivery. However, Fu-Jian Xu's research team reported a PBA-hybrid disulfide polyaminoglycoside polymer (SS-HPT-P, Fig. 4) for the delivery of CRISPR-Cas9. The SS-HPT-P method can be used to transfect CRISPR-Cas9 plasmids in A549 cell lines. SS-HPT-P/pCas9-surv has demonstrated effective tumor targeting in both in vivo and in vitro settings, reducing sensitive degradability.87In general, interaction between hydroxyl groups of a sugar and boronate cis-diol typically occurs. Mangesh Morey and his team have developed a glucose-responsive submicron particle (SMP) system for the efficient delivery of NIH3T3 cell genes.88 The research team from the Institute for Basic Science in Korea successfully constructed a dual stimuli-responsive vehicle for gene payloads and release, without any negative effects on cell viability.89
Polyanionic DNA showed strong interaction with cationic polymer structures like PBA, PEI, and PEG, influenced by an acidic endosomal pH environment. The Kaiyong Cai group developed hollow silica nanoparticles modified with PBA for dual-responsive delivery of doxorubicin in targeted tumor therapy.90 Li Chen and colleagues have published research on BA cross-linked dextran-PLA micelles or CNPBA-Dex-b-PLA for effective intracellular drug delivery into tumors. CNPBA-Dex-b-PLA exhibits exceptional pH sensitivity, indicating the optimal conditions for medication release from micelles. The MTT assay revealed that CNPBA-Dex-b-PLA, a drug delivery system, demonstrated good cytocompatibility with cell viability exceeding 90%.91 Bin Yang et al. developed a self-assembled PEI1.8K–boronic acid–cyclodextrin complex system to deliver pDNA and the anticancer drug doxorubicin (DOX). Enhancing drug efficacy for improved cancer treatment could be possible with this bio-responsive co-delivery nanotechnology.92
Ronald T. Raines et al. successfully delivered a hydrophilic macromolecule into human cells using boronate as a noncationic vector, combining BA with saccharides.93 Consequently, Zhao et al. created a parallel block copolymer micelle for healthy cells, which improved uptake and reduced cytotoxicity in in vitro uptake and cytotoxicity assays. Therefore, this straightforward decorating technique may make PBA-targeted tumor growth easier.94 Jihui Yu's research group developed a new approach to increase therapeutic efficacy of camptothecin using a combination of PBA, polyglutamic acid, PEG, and disulfide-bonded CPT.95 PBA-PEG-P (Glu-co-GlussCPT) demonstrated antiproliferative action against tumor cells, improved endocytosis efficiency, and notable in vivo anticancer activity in murine and human hepatoma xenograft models without systemic toxicity. Yiffan Ma and his research team developed a nanovector for cancer-targeted siRNA delivery using PBA and low molecular weight PEI 1.8K (PEI-PBA), which showed excellent cancer-targeted RNA delivery by recognizing the SA present on cancer cell membranes, a high level of biocompatibility and RNase resistance, and serum stability.96 The study synthesized elastin-like polypeptide (ELP) nanoparticles by polymerizing N-3-acrylamidophenylboronic acid (APBA) in the presence of ELP, which was expressed from a plasmid in Escherichia coli. ELP-PAPBA (Fig. 4), nanoparticles that are incorporated with PBA, have a spherical shape and are highly stable in an aqueous medium with a wide pH range. This clearly shows that the BA group plays a significant role in the internalization of ELP-PAPBA nanoparticles as evidenced by the ease with which these ELP-PAPBA nanoparticles were taken up by SH-SY5Y cancer cells and the ability of free APBA and SA to prevent this uptake.97 Polyplex micelles were created using siRNAs-3-fluoro-4-carboxyphenylboronic acid (FPBA)-adapted PEG-b-Plys, which contains esters from PBA and 1,2- or 1,3-cis-diols on a ribose ring (Fig. 4). Polyplex micelles, incorporating three envoy approaches, successfully stabilize PIC-based siRNA vectors while maintaining environmental sensitivity, and were successfully used for siRNA delivery in the OSRC-2 human renal carcinoma cell line.97 Experimental studies have shown that pendant BAs facilitate the successful delivery of RNase A into the cytosol.93
Huayue Wu's research team developed a dual responsive hydrogel, combining low molecular weight hydrogel-oligopeptide and PBA, for drug delivery, sugar/pH detection, and controlled release. In vitro experimental studies revealed that hydrogels are capable of sugar/pH detection and self-tuning controlled-release, and can be used in treating diseases like diabetes and cancer.98 The research group from the University of Washington, developed BA-containing copolymers for direct loading, which could be trafficked into acidic compartments and showed minimal cytotoxicity. Moreover, this polymer–drug conjugate effectively delivered Bis-T-23 into cultured podocytes.99
The prognosis of hepatocellular carcinoma (HCC) is poor due to its diagnosis in the unresectable phase and limited treatment options. Polymerized PBA–doxorubicin hybrid nanoparticles, traditionally used in HCC treatment, have been generated to enhance cancer healing and anticancer effects. The nanoparticles demonstrated excellent anti-tumor activity in subcutaneous and orthotopic HCC syngeneic mouse tumor models, suggesting targeted DOX delivery to liver cancer, where SA is overexpressed.100 A Kansas State University research group developed polymeric nanoparticles loaded with curcumin to address its rapid degradation and minimal solubility in water. The conjugation of PBA with curcumin in polymeric nanoparticles enhanced the chemical stability of curcumin and its sustained release under physical conditions. Furthermore, polymeric nanoparticles significantly increased the antiangiogenic and anticancer activities.101 PBA-CS-VE-decorated nanomaterials demonstrated strong mucin adhesion, minimal cytotoxicity, biocompatibility, and maximum cellular uptake in HCE-T-cell studies.102
6. Conclusion
Nanoparticles of phenylboronic acid (PBA) and its derivatives have tremendous promise for targeted delivery and advanced diagnostics. Their distinct sugar-binding capabilities, customizable qualities, and continuous development make them an adaptable platform for investigating innovative therapeutic and diagnostic techniques. In this review, we have presented an overview of the synergistic benefits of combining the use of PBA nanoparticles in various treatment methods. We also described the role of PBA-conjugated macromolecules in drug/gene delivery. The chemistry of PBA's binding with and dissociation from cell-surface glycans, as well as the implications for drug administration, have been discussed with examples. Future drug delivery research in this area requires more advancements that combine PBA with other functional entities, such as imaging agents or stimuli-responsive components. This multifunctionality may enable simultaneous diagnosis and therapy or improve therapeutic effectiveness.Conflicts of interest
Regarding the publishing of this paper, the authors state that they have no conflicts of interest.Acknowledgements
Dr Venkanna Muripiti thanks the vice chancellor of the Central University of Kerala for his constant support and encouragement. Dr Janardhan Banothu expresses his gratitude to DST-SERB for the research grant (Grant No: EEQ/2020/000303) and thanks the Director of NIT Calicut for providing the research facilities.References
- N. Rivlin, R. Brosh, M. Oren and V. Rotter, Genes Cancer, 2011, 2, 466–474 CrossRef CAS PubMed.
- Y. Xiong, M. Li, Q. Lu, G. Qing and T. Sun, Polymers, 2017, 9, 249 CrossRef PubMed.
- G. A. Rao and D. D. Buethe, J. Drug Targeting, 2004, 12, 341–345 CrossRef CAS PubMed.
- S. J. Henley, C. C. Thomas, D. R. Lewis, E. M. Ward, F. Islami, M. Wu, H. K. Weir, S. Scott, R. L. Sherman, J. Ma, B. A. Kohler, K. Cronin, A. Jemal, V. B. Benard and L. C. Richardson, Cancer, 2020, 126, 2250–2266 CrossRef PubMed.
- B. Mansoori, A. Mohammadi, S. Davudian, S. Shirjang and B. Baradaran, Adv. Pharm. Bull., 2017, 7, 339–348 CrossRef CAS PubMed.
- Y. Zhang, M. Li, X. Gao, Y. Chen and T. Liu, J. Hematol. Oncol., 2019, 12, 137 CrossRef PubMed.
- P. Kesharwani, R. Chadar, A. Sheikh, W. Y. Rizg and A. Y. Safhi, Front. Pharmacol., 2022, 12, 800481 CrossRef PubMed.
- Z. Wang, K. Zhao, T. Hackert and M. Zöller, Front. Cell Dev. Biol., 2018, 6, 97 CrossRef PubMed.
- P. Charkhchi, C. Cybulski, J. Gronwald, F. O. Wong, S. A. Narod and M. R. Akbari, Cancers, 2020, 12, 3730 CrossRef CAS PubMed.
- W. Chen, Z. Zhang, S. Zhang, P. Zhu, J. K.-S. Ko and K. K.-L. Yung, Int. J. Mol. Sci., 2021, 22, 6567 CrossRef CAS PubMed.
- C. Kratochwil, F. L. Giesel, M. Stefanova, M. Benešová, M. Bronzel, A. Afshar-Oromieh, W. Mier, M. Eder, K. Kopka and U. Haberkorn, J. Nucl. Med., 2016, 57, 1170–1176 CrossRef CAS PubMed.
- C. Dobie and D. Skropeta, Br. J. Cancer, 2021, 124, 76–90 CrossRef CAS PubMed.
- A. Y. Berghuis, J. F. A. Pijnenborg, T. J. Boltje and J. M. A. Pijnenborg, Int. J. Cancer, 2022, 150, 678–687 CrossRef CAS PubMed.
- C. O. Soares, A. S. Grosso, J. Ereno-Orbea, H. Coelho and F. Marcelo, Front. Mol. Biosci., 2021, 8, 727847 CrossRef CAS PubMed.
- R. Hombu, S. Neelamegham and S. Park, Molecules, 2021, 26, 5950 CrossRef CAS PubMed.
- D. Munoz-Provencio and M. J. Yebra, Int. J. Mol. Sci., 2023, 24, 9994 CrossRef CAS PubMed.
- M. Mullerova, M. Hovorkova, T. Zavodna, L. C. Stastna, A. Krupkova, V. Hamala, K. Novakova, J. Topinka, P. Bojarova and T. Strasak, Biomacromolecules, 2023, 24, 4705–4717 CrossRef CAS PubMed.
- B. Ramakrishnan and P. K. Qasba, J. Mol. Biol., 2007, 365, 570–576 CrossRef CAS PubMed.
- A. Varki, Trends Mol. Med., 2008, 14, 351–360 CrossRef CAS PubMed.
- P. Burzynska, L. F. Sobala, K. Mikolajczyk, M. Jodlowska and E. Jaskiewicz, Biomolecules, 2021, 11, 831 CrossRef CAS PubMed.
- T. Angata and A. Varki, Chem. Rev., 2002, 102, 439–469 CrossRef CAS PubMed.
- A. Varki, Nature, 2007, 446, 1023–1029 CrossRef CAS PubMed.
- D. Ilver, P. Johansson, H. Miller-Podraza, P. G. Nyholm, S. Teneberg and K. A. Karlsson, Methods Enzymol., 2003, 363, 134–157 CAS.
- F. Lehmann, E. Tiralongo and J. Tiralongo, Cell. Mol. Life Sci., 2006, 63, 1331–1354 CrossRef CAS PubMed.
- E. R. Vimr, K. A. Kalivoda, E. L. Deszo and S. M. Steenbergen, MMBR, Microbiol. Mol. Biol. Rev., 2004, 68, 132–153 CrossRef CAS PubMed.
- H. E. Hendin, P.-O. Lavoie, J. M. Gravett, S. Pillet, P. Saxena, N. Landry, M.-A. D'Aoust and B. J. Ward, npj Vaccines, 2022, 7, 42 CrossRef CAS PubMed.
- X. Zhou, G. Yang and F. Guan, Cells, 2020, 9(2), 273 CrossRef CAS PubMed.
- S. Hakomori, Cancer Res., 1996, 56, 5309–5318 CAS.
- D. H. Dube and C. R. Bertozzi, Nat. Rev. Drug Discovery, 2005, 4, 477–488 CrossRef CAS PubMed.
- L. Daniel, J. Trouillas, W. Renaud, P. Chevallier, J. Gouvernet, G. Rougon and D. Figarella-Branger, Cancer Res., 2000, 60, 80–85 CAS.
- C. Dobie and D. Skropeta, Br. J. Cancer, 2020, 124, 76–90 CrossRef PubMed.
- L. A. Liotta and W. G. Stetler-Stevenson, Cancer Res., 1991, 51, 5054s–5059s CAS.
- V. Muripiti, L. Brijesh, H. K. Rachamalla, S. K. Marepally, R. Banerjee and S. V. Patri, Bioorg. Chem., 2019, 82, 178–191 CrossRef CAS PubMed.
- V. Muripiti, H. K. Rachamalla, R. Banerjee and S. V. Patri, Org. Biomol. Chem., 2018, 16, 2932–2946 RSC.
- V. Muripiti, T. Y. Mujahid, V. H. V. Boddeda, S. Tiwari, S. K. Marepally, S. V. Patri and V. Gopal, Int. J. Pharm., 2019, 554, 134–148 CrossRef CAS PubMed.
- G. Zhang, J. Gao, J. Qian, L. Zhang, K. Zheng, K. Zhong, D. Cai, X. Zhang and Z. Wu, ACS Appl. Mater. Interfaces, 2015, 7, 14192–14200 CrossRef CAS PubMed.
- L. Liu, X. Ma, Y. Chang, H. Guo and W. Wang, Biosensors, 2023, 13, 785 CrossRef CAS PubMed.
- M. P. Silva, L. Saraiva, M. Pinto and M. E. Sousa, Molecules, 2020, 25, 4323 CrossRef CAS PubMed.
- H. J. Lo, L. Krasnova, S. Dey, T. Cheng, H. Liu, T. I. Tsai, K. B. Wu, C. Y. Wu and C. H. Wong, J. Am. Chem. Soc., 2019, 141, 6484–6488 CrossRef CAS PubMed.
- A. Matsumoto and Y. Miyahara, Sci. Technol. Adv. Mater., 2018, 19, 18–30 CrossRef CAS PubMed.
- R. Smoum and M. Srebnik, in Contemporary Aspects of Boron: Chemistry and Biological Applications, 2005, vol. 22, pp. 391–494 Search PubMed.
- B. J. Graham, I. W. Windsor, B. Gold and R. T. Raines, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2013691118 CrossRef CAS PubMed.
- B. Akgun and D. G. Hall, Angew. Chem., Int. Ed. Engl., 2018, 57, 13028–13044 CrossRef CAS PubMed.
- G. F. Whyte, R. Vilar and R. Woscholski, J. Chem. Biol., 2013, 6, 161–174 CrossRef PubMed.
- H. Laubli and L. Borsig, Front. Immunol., 2019, 10, 2120 CrossRef CAS PubMed.
- S. Senapati, A. K. Mahanta, S. Kumar and P. Maiti, Signal Transduction Targeted Ther., 2018, 3, 7 CrossRef PubMed.
- S. Deshayes, H. Cabral, T. Ishii, Y. Miura, S. Kobayashi, T. Yamashita, A. Matsumoto, Y. Miyahara, N. Nishiyama and K. Kataoka, J. Am. Chem. Soc., 2013, 135, 15501–15507 CrossRef CAS PubMed.
- R. R. Castillo, D. Lozano, B. Gonzalez, M. Manzano, I. Izquierdo-Barba and M. Vallet-Regi, Expert Opin. Drug Delivery, 2019, 16, 415–439 CrossRef CAS PubMed.
- W. L. A. Brooks, C. C. Deng and B. S. Sumerlin, ACS Omega, 2018, 3, 17863–17870 CrossRef CAS PubMed.
- M. Melendez, O. Rosario, B. Zayas and J. F. Rodriguez, J. Pharm. Biomed. Anal., 2009, 49, 1233–1240 CrossRef CAS PubMed.
- X. Chen, S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler and F. M. Hughson, Nature, 2002, 415, 545–549 CrossRef CAS PubMed.
- G. Pasparakis, A. Cockayne and C. Alexander, J. Am. Chem. Soc., 2007, 129, 11014–11015 CrossRef CAS PubMed.
- E. Uchimura, H. Otsuka, T. Okano, Y. Sakurai and K. Kataoka, Biotechnol. Bioeng., 2001, 72, 307–314 CrossRef CAS PubMed.
- S. Jin, Y. Cheng, S. Reid, M. Li and B. Wang, Med. Res. Rev., 2010, 30, 171–257 CrossRef CAS PubMed.
- J. P. M. Antonio, R. Russo, C. P. Carvalho, P. Cal and P. M. P. Gois, Chem. Soc. Rev., 2019, 48, 3513–3536 RSC.
- M. Z. H. Kazmi, J. P. G. Rygus, H. T. Ang, M. Paladino, M. A. Johnson, M. J. Ferguson and D. G. Hall, J. Am. Chem. Soc., 2021, 143, 10143–10156 CrossRef CAS PubMed.
- S. Chatterjee, E. V. Anslyn and A. Bandyopadhyay, Chem. Sci., 2020, 12, 1585–1599 RSC.
- Y. Y. Aung, A. N. Kristanti, H. V. Lee and M. Z. Fahmi, ACS Omega, 2021, 6, 17750–17765 CrossRef CAS PubMed.
- R. Melavanki, R. Kusanur, K. K. Sadasivuni, D. Singh and N. R. Patil, Heliyon, 2020, 6, e05081 CrossRef CAS PubMed.
- J. Wang, W. Wu, Y. Zhang, X. Wang, H. Qian, B. Liu and X. Jiang, Biomaterials, 2014, 35, 866–878 CrossRef CAS PubMed.
- B. Marco-Dufort, R. Iten and M. W. Tibbitt, J. Am. Chem. Soc., 2020, 142, 15371–15385 CrossRef CAS PubMed.
- N. A. Siddiqui, N. Billa, C. J. Roberts and Y. A. Osei, Pharmaceutics, 2016, 8, 30 CrossRef PubMed.
- X. Xue, H. Gong, H. Zheng and L. Ye, ACS Appl. Nano Mater., 2021, 4, 2866–2875 CrossRef CAS PubMed.
- Y. Yao, Y. Zhou, L. Liu, Y. Xu, Q. Chen, Y. Wang, S. Wu, Y. Deng, J. Zhang and A. Shao, Front. Mol. Biosci., 2020, 7, 193 CrossRef CAS PubMed.
- P. N. Navya, A. Kaphle, S. P. Srinivas, S. K. Bhargava, V. M. Rotello and H. K. Daima, Nano Convergence, 2019, 6, 23 CrossRef CAS PubMed.
- A. Wicki, D. Witzigmann, V. Balasubramanian and J. Huwyler, J. Controlled Release, 2015, 200, 138–157 CrossRef CAS PubMed.
- S. G. Crich, D. Alberti, I. Szabo, S. Aime and K. Djanashvili, Angew. Chem., Int. Ed. Engl., 2013, 52, 1161–1164 CrossRef PubMed.
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nat. Nanotechnol., 2007, 2, 751–760 CrossRef CAS PubMed.
- V. Rotello, Cell Cycle, 2009, 8, 3615–3616 CrossRef CAS PubMed.
- Q. Tang, J. Liu, Y. Jiang, M. Zhang, L. Mao and M. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 46585–46590 CrossRef CAS PubMed.
- R. Wang, C. Yin, C. Liu, Y. Sun, P. Xiao, J. Li, S. Yang, W. Wu and X. Jiang, J. Am. Chem. Soc., 2021, 143, 20927–20938 CrossRef CAS PubMed.
- W. J. Kim, Y. J. Kwon, C. H. Cho, S. K. Ye and K. O. Kim, Sci. Rep., 2021, 11, 21894 CrossRef CAS PubMed.
- M. Akbarian, A. Khani, S. Eghbalpour and V. N. Uversky, Int. J. Mol. Sci., 2022, 23, 1445 CrossRef CAS PubMed.
- A. Gross, C. Hashimoto, H. Sticht and J. Eichler, Front. Bioeng. Biotechnol., 2015, 3, 211 Search PubMed.
- L. Wang, N. Wang, W. Zhang, X. Cheng, Z. Yan, G. Shao, X. Wang, R. Wang and C. Fu, Signal Transduction Targeted Ther., 2022, 7, 48 CrossRef CAS PubMed.
- T. Luo, W. Huang, F. Chu, T. Zhu, B. Feng, S. Huang, J. Hou, L. Zhu, S. Zhu and W. Zeng, Mol. Pharm., 2023, 20, 4942–4970 CrossRef CAS PubMed.
- R. Smoum, A. Rubinstein, V. M. Dembitsky and M. Srebnik, Chem. Rev., 2012, 112, 4156–4220 CrossRef CAS PubMed.
- H. S. Ban and H. Nakamura, Chem. Rec., 2015, 15, 616–635 CrossRef CAS PubMed.
- A. R. Martin, J. J. Vasseur and M. Smietana, Chem. Soc. Rev., 2013, 42, 5684–5713 RSC.
- J. Whited, C. K. Rama and X. L. Sun, ACS Omega, 2018, 3, 13467–13473 CrossRef CAS PubMed.
- W. Zhang, D. I. Bryson, J. B. Crumpton, J. Wynn and W. L. Santos, Chem. Commun., 2013, 49, 2436–2438 RSC.
- J. E. Wynn, W. Zhang, D. M. Tebit, L. R. Gray, M. L. Hammarskjold, D. Rekosh and W. L. Santos, Bioorg. Med. Chem., 2016, 24, 3947–3952 CrossRef CAS PubMed.
- Y. Sumie, K. Sato, T. Kakegawa and Y. Furukawa, Commun. Chem., 2023, 6, 89 CrossRef CAS PubMed.
- D. Wu, J. Yang, Z. Xing, H. Han, T. Wang, A. Zhang, Y. Yang and Q. Li, Colloids Surf., B, 2016, 146, 318–325 CrossRef CAS PubMed.
- Y. Liang, Y. Wang, L. Wang, Z. Liang, D. Li, X. Xu, Y. Chen, X. Yang, H. Zhang and H. Niu, Bioact. Mater., 2021, 6, 433–446 CAS.
- C. Liu, T. Wan, H. Wang, S. Zhang, Y. Ping and Y. Cheng, Sci. Adv., 2019, 5, eaaw8922 CrossRef CAS PubMed.
- M. Shao, Y. Qi, D. Sui and F. J. Xu, Biomater. Sci., 2021, 9, 7104–7114 RSC.
- M. Morey, A. Srivastava and A. Pandit, Pharmaceutics, 2021, 13, 62 CrossRef CAS PubMed.
- J. Kim, Y. M. Lee, H. Kim, D. Park and W. J. Kim, Biomaterials, 2016, 75, 102–111 CrossRef CAS PubMed.
- L. Huang, Q. Zhang, L. Dai, X. Shen, W. Chen and K. Cai, Regener. Biomater., 2017, 4, 111–124 CAS.
- Z. Zhao, X. Yao, Z. Zhang, L. Chen, C. He and X. Chen, Macromol. Biosci., 2014, 14, 1609–1618 CrossRef CAS PubMed.
- B. Yang, H. Jia, X. Wang, S. Chen, X. Zhang, R. Zhuo and J. Feng, Adv. Healthcare Mater., 2014, 3, 596–608 CrossRef CAS PubMed.
- G. A. Ellis, M. J. Palte and R. T. Raines, J. Am. Chem. Soc., 2012, 134, 3631–3634 CrossRef CAS PubMed.
- D. Zhao, J. Q. Xu, X. Q. Yi, Q. Zhang, S. X. Cheng, R. X. Zhuo and F. Li, ACS Appl. Mater. Interfaces, 2016, 8, 14845–14854 CrossRef CAS PubMed.
- Y. Huang, W. Zhang, Y. Xu, S. Zhu, Y. Wu, T. Chen, Y. Xiao, W. Lu, X. Zhang and J. Yu, Int. J. Pharm., 2020, 580, 119250 CrossRef CAS PubMed.
- M. Ji, P. Li, N. Sheng, L. Liu, H. Pan, C. Wang, L. Cai and Y. Ma, ACS Appl. Mater. Interfaces, 2016, 8, 9565–9576 CrossRef CAS PubMed.
- M. Naito, T. Ishii, A. Matsumoto, K. Miyata, Y. Miyahara and K. Kataoka, Angew. Chem., Int. Ed., 2012, 51, 10751–10755 CrossRef CAS.
- N. Tao, G. Li, M. Liu, W. Gao and H. Wu, Tetrahedron, 2017, 73, 3173–3180 CrossRef CAS.
- Y. Cheng, G. W. Liu, R. Jain, J. W. Pippin, S. J. Shankland and S. H. Pun, ACS Biomater. Sci. Eng., 2018, 4, 3968–3973 CrossRef CAS.
- B. Y. Kang, W. Hur, S. M. Kim, S. Kim, J. Lee, E. Tak, W. J. Kim, H. J. Chun and S. K. Yoon, Nanomedicine, 2021, 34, 102389 CrossRef CAS.
- A. J. van der Vlies, M. Morisaki, H. I. Neng, E. M. Hansen and U. Hasegawa, Bioconjugate Chem., 2019, 30, 861–870 CrossRef CAS.
- X. Sun, Y. Sheng, K. Li, S. Sai, J. Feng, Y. Li, J. Zhang, J. Han and B. Tian, Acta Biomater., 2022, 138, 193–207 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2024 |