Open Access Article
Open Access ArticleAdvancements in amyloid-based biological materials for healthcare, environmental and sensing applications
Smriti Singh
Yadav
ab,
Prabeen Kumar
Padhy
 ab,
Ashish Kumar
Singh
ab,
Ashish Kumar
Singh
 c,
Supriya
Sharma
a,
Tanu
c,
Siraj
Fatima
ab,
Anurag
Sinha
bc,
Ramsha
Tariq
a,
Varsha
bc,
Sandeep K.
Sharma
c,
Supriya
Sharma
a,
Tanu
c,
Siraj
Fatima
ab,
Anurag
Sinha
bc,
Ramsha
Tariq
a,
Varsha
bc,
Sandeep K.
Sharma
 bc and
Smriti
Priya
bc and
Smriti
Priya
 *ab
*ab
aSystems Toxicology Group, FEST Division, CSIR-Indian Institute of Toxicology Research, Vishvigyan Bhawan, 31 Mahatma Gandhi Marg, Lucknow 226001, Uttar Pradesh, India. E-mail: spriya@iitr.res.in; smritipriya3@gmail.com
bAcademy of Scientific and Innovative Research (AcSIR), Ghaziabad 201002, India
cFood Toxicology Group, FEST Division, CSIR-Indian Institute of Toxicology Research, Vishvigyan Bhawan, 31 Mahatma Gandhi Marg, Lucknow 226001, Uttar Pradesh, India
First published on 30th April 2024
Abstract
Amyloids are insoluble accumulations of fibrils formed by various proteins such as Aβ1–40, lysozyme, α-synuclein, insulin and peptides associated with several protein aggregation-related diseases. The self-assembling property of amyloid fibrils has been demonstrated to be a powerful characteristic for creating protein-based materials for various applications. Biomaterials based on amyloid fibrils have recently gained momentum due to their superior mechanical strength, exceptional self-assembly and stability in various matrices. Owing to their versatile structural characteristics, amyloid fibrils can have remarkable potential for application in medicine, materials, packaging, tissue engineering, sensors and drug delivery. Here, the recent advancements in the structural characterization, synthesis, classification and the emerging applications of amyloid fibrils in healthcare, sensing and the environment have been reviewed. The biocompatibility, biodegradability and structural stability of amyloid fibril-based biomaterials open up new avenues for sustainable and eco-friendly alternatives to the conventionally used synthetic materials.
1. Introduction
Globally, industrial and consumer preferences are shifting towards green, sustainable and eco-friendly alternatives to conventionally used synthetic materials.1 Utilizing renewable agro-resources to develop biodegradable materials has gained significant attention. Various biopolymers are found in natural sources, such as proteins, starch, cellulose fibres and lipids.2,3 Proteins and peptides are functionally important biomolecules performing several functions in the maintenance of cellular homeostasis.4 Proteins have shown to provide the molecular basis for the development of various biomaterials.5 Natural protein-based biomaterials such as silk proteins and egg hen lysozyme demonstrate the ability of proteins to self-assemble and aggregate to form functional materials.6,7 The biocompatibility and structural characteristics of proteins and peptide assemblies have tremendous potential in biomaterial applications. Under physiological conditions, proteins fold to their native three-dimensional structure; however, exposure to various stress conditions they may misfold and aggregate due to non-native interactions and form amyloid fibrils.4,8 The formation of amyloid fibrils is related to proteins with amyloid cores and has a self-assembling and aggregation propensity.9 The amyloid protein aggregates were first identified as disease hallmarks associated with several protein aggregation-related diseases, including Alzheimer's and Parkinson's disease.10 Structurally, amyloid fibrils are composed of β-sheets formed by the structural transition of the helical structure and are categorized as supramolecular polymers of β-strands stacked perpendicularly to the fibrils on their long axis.11 Amyloids are insoluble accumulations of fibrils formed by various independent proteins such as Aβ1–40, α-syn, insulin and peptides.12,13 Apart from pathologically associated proteins such as amyloid β (Aβ1–40), tau and α-synuclein (α-syn), several other proteins also form amyloid fibrils at high concentrations and under destabilizing conditions.8,13 The protein aggregates and amyloids are formed when the soluble physiological native form of protein undergoes misfolding and forms a nucleus for aggregation that further elongates by the addition of misfolded monomers.14,15 The protein aggregates formed thus include small oligomeric precursors, amorphous aggregates and mature fibrils, each exhibiting distinct structural and morphological characteristics.16Several functional forms of amyloids have been identified and characterized in plants and animals, showing potential for diverse applications.17,18 The self-assembling property of amyloid fibrils has been demonstrated to be a powerful characteristic for creating protein-based materials for various applications. Functional amyloids have numerous applications in metal–organic frameworks (MOF) as sensors for amyloid diseases, whey protein isolate amyloid fibrils in combination with carboxymethyl cellulose as a drug delivery vehicle, membrane formation, nanosensor applications, and as membranes for packaging and healthcare applications.19–21 The biocompatibility, biodegradability and structural stability of amyloid fibrils can present a sustainable and eco-friendly alternative to the conventionally used synthetic materials.22 The stable proteinaceous biostructures formed by amyloid fibrils are structurally flexible, allowing modification using chemical modification, denaturation, enzymatic modification, nanomaterials and bioactive molecules to develop sensors, embedded nanomaterials and membranes.23,24
In the present review, the recent advancements in structural characterization, synthesis, classification, and the emerging applications of amyloid fibrils in healthcare, sensing and the environment have been reviewed. Further, an overview of the protein-based materials available from natural sources and their role in drug delivery and sensor applications has been provided. Biomaterials based on amyloid protein aggregates have several advantages due to their superior mechanical strength, exceptional self-assembly and stability in various matrices. The biocompatible and biodegradable nature of amyloid fibril-based biomaterials and their versatile structural properties can provide a sustainable and eco-friendly alternative to conventionally used synthetic materials.
2. Structural characteristics of amyloid fibrils
2.1 Formation of amyloid fibrils
The application of amyloid fibrils in developing functional materials and membranes has gained interest due to their extended, highly stable β-sheet structures and β-turns.25 Here, partially misfolded monomers result in the assembly of oligomers, ultimately leading to the formation of fibrils as shown in Fig. 1.26 The transition state of aggregation is represented by oligomer formation, where the metastable β-sheet conformations dominate and gradually result in the formation of β-sheet rich structures. The formation of oligomeric aggregates further leads to rapid nucleation of monomers to oligomers or fibrils.27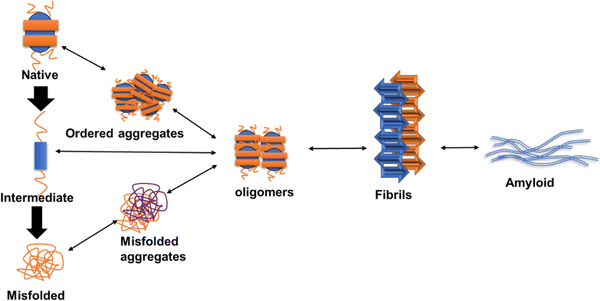 | ||
| Fig. 1 The process of amyloid formation is initiated with protein misfolding, oligomer formation and fibril accumulation leading to amyloid formation. | ||
The fundamental structure of amyloids was first identified in the 1950s with the advancement in electron microscopy techniques and X-ray fiber diffraction.28,29 The distinguishing elements of the fibrillar structure of amyloids are present at the different structural levels, such as tertiary and quaternary, which can be easily identified using nuclear magnetic rasonance (NMR), cryo electron microscopy (cryo-EM) and solid-state nuclear magnetic resonance (SSNMR).30 Amyloid fibrils have a needle-like and unbranched morphology consisting of several bundled protofilaments. Each protofilament comprises cross-β structure sheets that are parallel to the fibrillar axis and stacked perpendicular to the main axis.28 The fibrillar arrangement depends upon the exposure to different conditions such as agitation, salt concentration, protein concentration, temperature, pH, and the degree of folding and unfolding during fibril formation.31 Various peptides frequently assemble β-sheets by stacking elongated β-strands, and a steric zipper is formed when two β-sheets are strongly interdigitated with adjacent chains.32 The intricate structure and high organization of amyloid fibrils prevent their breakdown by proteases.33 These characteristics of amyloid fibrils are due to a dense backbone of hydrogen bonding network in cross-β structures, that provides stiffness up to the gigapascal (GPa) level.28 Various studies have also reported diversity in amyloid structure formation from a single protein, referred to as amyloid polymorphism. Depending on different fibril growth conditions such as temperature, solvents, protein concentrations and agitation, it results in the generation of amyloid polymorphs that are produced in vitro and are also related to protein aggregation diseases.28
2.2. Structural characterization of amyloid fibrils
Various biophysical approaches can be employed to characterize amyloids based on their structure and stability. Among these, X-ray diffraction is one of the standard and classical methods that investigates the formation of cross-β structures.34 The widely used method to determine the structure and track the formation of amyloid aggregation is the thioflavin T (ThT) assay, which binds specifically to the β-sheet structures formed in the amyloids.35 Various other biophysical methods are used to determine the presence of β-sheet rich structures in amyloid fibrils, such as Fourier transform infrared (FTIR) spectroscopy and circular dichroism (CD) for analyzing secondary structures, transmission electron microscopy (TEM), scanning electron microscopy (SEM) and atomic force microscopy (AFM) for structure visualization, and super-resolution microscopy (SRM) and light/neutron/X-ray scattering for microscopic analysis of amyloid structures.36 The molecular and atomic scale investigation of the amyloid morphology and structural studies done by hydrogen–deuterium (H/D) exchange and cryo-EM techniques are alternatives to conventional techniques such as X-ray crystallography and NMR.37 Other methods, such as dynamic light scattering (DLS) and quartz crystal microbalance (QCM), are used to monitor misfolding, fibril elongation and aggregation.38 Characterization of protein-based membranes by X-ray diffraction reveals that the membranes possess a high level of ordered organization ranging from the nano to micrometer scale. X-ray diffraction can analyze the structure specific diffraction patterns, providing information about individual β-strands and protein nanofibrils.39 As a single biophysical technique cannot provide enough information on the detailed structure of amyloid fibrils, multiple techniques are employed to minimize the possibility of artefacts. Among the diverse techniques available, the most used techniques for characterizing amyloid fibrils are explained below.3. Amyloid-based materials
Highly stable amyloid fibrils are currently being used to develop biocompatible and biodegradable materials. These properties have increased scope for the development of functional amyloid materials such as hydrogels, scaffolds, membranes and nanomaterials for application in therapeutics, electronics, food packaging and the environment.46 Amyloid fibrils are assembled and modified to produce amyloid membranes that can be utilized for water purification, conductive three-dimensional aerogel biomaterials with sensing properties and drug-releasing vehicles.20,47 The amyloid protein-based membranes are being developed as hybrid materials with desired functional properties for use in drug delivery, water purification and nanofiltration. Hybrid amyloid membranes prepared from amyloids of β-lactoglobulin and porous carbon hybrids are used for removing arsenic contamination from water.48 Further, modified protein amyloid fibril/zirconium oxide (ZrO2) nanoparticles are utilized for removing fluorides from water.49 A hybrid membrane synthesised from carboxymethyl cellulose (CMC) and whey protein isolate amyloid fibril (WPI-AF) via chemical crosslinking coupled with phase inversion has been used as a drug delivery vehicle for methylene blue (MB) and riboflavin.20 Similarly, graphene oxide (GO)/amyloid-based membranes are also being used in water purification.50 The detailed applications of various amyloid-based materials are discussed in Sections 5.1–5.3.4. Natural protein-based materials
4.1. Concept of protein membranes
Proteins exist in various distinct structural conformations, such as monomers, small oligomers (homo/hetero-compositions), elongated fibrils and rods. Structural conformers of proteins such as lysozyme and β-lactoglobulin amyloid fibrils undergo polymerization, resulting in the formation of membranes and functional materials.51 The multi-level organisation of the protein structure allows interaction and intermolecular binding with different amino acids resulting in the formation of several linkages.23 Protein structure and stability modified using methods such as denaturation, chemical hydrolysis and crosslinking provides strength and reduces flexibility and permeability to different gases, vapours and liquids, required for membrane formation.23,52 Efforts are being made for environment-friendly approaches to create sustainable packaging materials from naturally occurring biopolymers.53 The application of membranes developed from proteins is being explored in food packaging to enhance the shelf life of food commodities.54 Protein-based membranes that are either animal- or plant-derived are becoming popular as compared to synthetic biopolymers due to their relative abundance in nature, ability to form membranes, biodegradability and nutritive value.55 Protein-based membranes have better gas and aroma-barrier properties along with the desired flexibility and mechanical strength as compared to lipid- and polysaccharide-derived membranes. Proteins isolated from animal sources such as collagen, gelatin, chitosan, and whey, and proteins from plant sources such as starch, gluten, zein, and gelatin are being used for the preparation of membranes.56–614.2. Major sources of proteins for biomaterial synthesis
Proteins, based on their structure, are classified into two categories: fibrous and globular proteins. Fibrous proteins such as collagen are water-insoluble and stabilized by intermolecular hydrogen bonds, whereas globular proteins such as wheat gluten, corn zein and soy proteins are water-soluble, and their structure is maintained by covalent (disulfide) bonds, hydrogen bonds, ionic and hydrophobic interactions.62 Their high abundance in nature and ease of processing, along with structural complexity and a higher tendency for intramolecular interactions, help in providing better mechanical properties, making proteins a suitable candidate for the synthesis of biomaterials.61,63 Therefore, several animal and plant proteins such as collagen, wheat gluten, corn zein, and soy proteins are highly used in the preparation of edible membranes,61 where the properties of the developed biomaterial or membrane depend on the nature of the protein, degree of chain extension and the sequence of amino acids. The developed biomaterials have desired flexibility, gas and liquid permeability, mechanical strength, shelf life and opacity,60 which can be enhanced using various additives such as plasticizers, vitamin E, nisin and cellulose.23,64 Thus, protein-based biomaterials are promising alternatives to synthetic packaging materials, and the natural sources of proteins utilized for the development of various functional materials are discussed in the following sections.4.3. Natural sources of protein membranes
Biomolecules such as proteins, carbohydrates and lipids have the potential to produce multifaceted biomaterials and membranes owing to their biodegradability and sustainability.65,66 Various animal and plant proteins such as soy protein, milk whey, casein, wheat proteins, corn zein, collagen, silk and oilseed proteins from different natural sources have been explored and studied for membrane production as listed in Tables 1 and 2.67 These biodegradable protein-based materials are used in medicine, biotechnology and packaging applications.68 Protein-based membranes and biomaterials are strong; however, non-plasticized membranes are brittle.69,70 To address this issue, various chemical and physical modifications of protein components are required during membrane production to enhance the mechanical properties and functional applications.71| Protein group | Plant source | Features of the membrane | Applications | |||
|---|---|---|---|---|---|---|
| Mechanical strength | Water vapour permeability | Thermal stability | Antioxidant activity | |||
| Legume proteins | Kidney beans81 | Enhanced | Not reported | Not reported | Not reported | Antimicrobial food packaging |
| Mung bean82 | Enhanced | Enhanced | Not reported | Enhanced | Edible coating | |
| Faba bean83 | Enhanced | Low | Enhanced | Not reported | Biodegradable packaging | |
| Soy bean84 | Enhanced | Low | Enhanced | Not reported | Bio-based packaging | |
| Horse gram85 | Enhanced | Enhanced | Not reported | Not reported | Antimicrobial food packaging, biomedical materials | |
| Fenugreek86 | Enhanced | Low | Moderate | Not reported | Bio-based packaging | |
| Red bean87 | Enhanced | Low | Not reported | Not reported | Packaging materials | |
| Nut and oilseed proteins | Peanut88 | Low | Low | Not reported | Enhanced | Antimicrobial food packaging |
| Sesame89 | Enhanced | Low | Enhanced | Not reported | Food packaging | |
| Canola90 | Enhanced | Not reported | Enhanced | Not reported | Food packaging | |
| Sunflower91 | Enhanced | Low | Not reported | Enhanced | Fresh fruit packaging | |
| Flax seed92 | Enhanced | Low | Enhanced | Not reported | Antimicrobial food packaging film | |
| Rape seed93 | Enhanced | Not reported | Not reported | Not reported | Edible food coating | |
| Castor bean94 | Enhanced | No change | Not reported | Not reported | Seedling packaging bags | |
| Sorghum95 | Enhanced | Enhanced | Not reported | Enhanced | Antimicrobial food packaging | |
| Cereal grain proteins | Wheat96 | Enhanced | Low | Enhanced | Not reported | UV-resistant edible coating |
| Oat97 | Enhanced | Low | Enhanced | Not reported | Biodegradable packaging | |
| Corn98 | Enhanced | Not reported | Enhanced | Not reported | Scaffolds, biomaterials | |
| Rice99 | Enhanced | Low | Enhanced | Enhanced | Antimicrobial food packaging | |
| Barley100 | Enhanced | Low | Enhanced | Not reported | Biodegradable packaging | |
| Pseudo-cereal proteins | Quinoa101 | Enhanced | Low | Not reported | Enhanced | Antimicrobial active packaging |
| Amaranth102 | Enhanced | Low | Not reported | Not reported | Edible food coating | |
| Source | Protein | Advantages | Limitations | Applications |
|---|---|---|---|---|
| Animal meat | Gelatin105–108 | Gas barrier properties, antioxidant and anti-bacterial activity | Weak water vapor barrier properties, moderate mechanical properties | Active food packaging, edible coating |
| Animal meat | Collagen109,110 | Antioxidant and anti-bacterial properties, barrier for moisture, high mechanical strength, thermostability | High water vapour transmission rate | Active edible food packaging |
| Animal meat | Myofibrillar protein111 | High tensile strength | Highly sensitive to water vapour | As a plasticizer in packaging |
| Milk protein | Caseins112–114 | Low oxygen permeability, moderate mechanical strength | Highly sensitive to water vapour, low elasticity | Packaging for cheese and food products |
| Milk protein | Whey protein115 | High mechanical strength, improved barrier properties, antibacterial and antioxidant properties | High vapour permeability, low tensile strength | Packaging |
4.4. Plant-based sources for biomaterials
Plant-based proteins derived from peas, beans, kernels, legumes and agro-waste have been used for preparing membranes and coatings.67,72–74 The presence of various amino acids and active binding sites in proteins helps to form molecular networks, providing desired elasticity or plasticity for preparing biopolymer membranes.75 Plant protein-based biopolymer membranes have issues with limited elongation properties, leading to brittleness in the developed materials and coatings.76 The structural characteristics and functional groups of the protein are exploited for enzymatic, chemical and physical modifications.77 Such modifications enhance their intrinsic barrier, plasticity, antimicrobial, antioxidant, thermal, mechanical and other biophysical properties.75,78,79 The use of plasticizers (e.g., glycerol, fatty acids and nanocomposites) significantly improves the functionality and bioactivity of the membranes.80 Various sources of plant protein-based materials and their applications are listed in Table 1.4.5. Animal-based sources
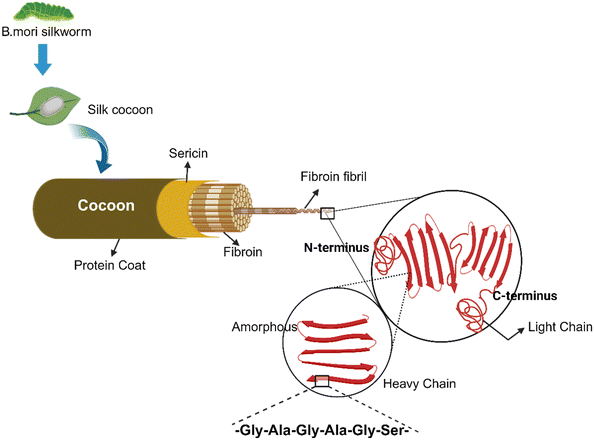
Various animal-derived biomolecules such as keratin, gelatin, collagen, silk fibroin/sericin and myofibrils have potential for synthesis of value-added biopolymer-based materials. Biopolymers derived from the waste by-products generated at slaughterhouses and meat processing industries can also be repurposed for developing biomaterials.103,104 Some of the widely used animal protein-based materials are described in Table 2.Sponges. Silk fibroin sponges are macroporous biomaterials used in tissue engineering. These porous sponges are prepared by mixing the silk solution through freeze-drying, gas foaming and addition of suitable porogens (polymers, sugar, salt crystals) through gelation.142
3D printed structures. Silk-based bio-inks and filaments are used as structural materials for the 3D bioprinting of custom geometric structures. Employing different 3D printing methods, complex and difficult-to-design customized biological constructs can be fabricated for biomedical applications.143
Electrospun mats. Electrospinning technology is used to create non-woven mats by spinning a polymeric solution in an electric field. Silk fibroins are used to produce nanofibrous mats by electrospinning the polymer solution through a needle and an electric force. These electrospun nanomats are used in grafts, implants, tissue scaffolds and other crucial healthcare applications.138,144
Silk-based hydrogels. Silk-based hydrogels and scaffolds are developed through chemical modifications via cross-linking of silk proteins for application in wound healing and medication.145 Hydrogels prepared using sericin–sodium alginate and silver nanoparticles (AgNPs) can potentially help in preventing bacterial infection.146 Similarly, hydrogels prepared by incorporating lysozyme, sericin and agarose are used to enhance antibacterial and water absorption properties.147 Sericin hydrogels prepared by crosslinking of glutaraldehyde are photoluminescent, enable cell adhesion, migration, and proliferation, and also help in long-term survival.148 Sericin and genipin cross-linked hydrogels have potential applications in the treatment of ischemic stroke and ischemic myocardial infarction,149 and also facilitate in vitro neuron attachment and neuroprotection.150
5. Application of amyloid-based functional materials
5.1. Amyloid-based materials in therapeutics and health
Amyloid fibrils have distinct features of self-assembly, structural stability and mechanical stiffness, making them promising candidates for healthcare interventions.151,152In vitro studies have revealed that amyloid fibrils derived from milk protein and plants are non-toxic towards Caco-2 and Hec-1a cell lines.153 Studies also reported an enhancement in the anti-inflammatory activity and bio-adhesivity of skin drugs used for topical treatments utilizing amyloid fibrils derived from milk protein and β-lactoglobulin.154 Amyloid-based hydrogels are dynamic structures and biocompatible, allowing fine-tuning of drug release in a controlled manner with high efficiency.155 A lysozyme-based amyloid microgel has been developed with potential drug-carrying and delivery properties through amyloid fibril networks,156 where ThT, Remazol Brilliant Blue R (RBBR), tetracycline and penicillin V have been demonstrated for their controlled release.157 Due to the physical cross-linking of amyloid-fibrils, hydrogels are formed that possess water-holding capability, biocompatibility and resilience to chemical and physical alterations while preserving their structural integrity.158,159 Injectable hydrogels have gained attention for administering various therapeutic molecules to the target organs owing to their compatibility with hydrophobic and hydrophilic drug molecules, peptides and other therapeutic compounds for targeted delivery as shown in Fig. 3.159 An example of such a system is the amyloid hydrogel derived from hen egg peptides, where an injectable drug carrier hydrogel synthesized using amyloid fibrils and peptide fragments obtained from hen egg white lysozyme were used.159 Doxorubicin, a chemotherapeutic agent, was used as a model compound to show the effectiveness of the developed injectable drug carrier system. The amyloid hydrogel exhibited low cytotoxicity and thixotropic properties.159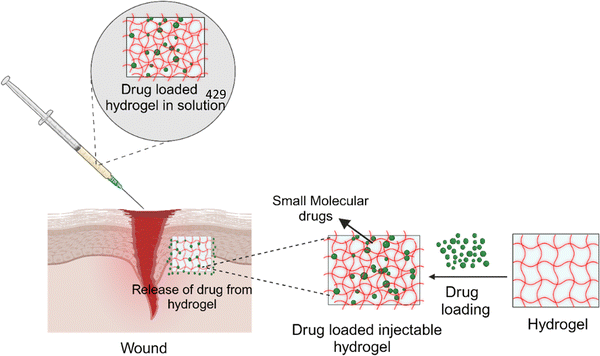 | ||
| Fig. 3 Schematic representation of amyloid-based hydrogels in drug loading and administration (created with BioRender.com). | ||
5.2. Amyloid-based materials
Amyloid-based materials are being widely used in medical applications such as tissue repair, scaffolds for cell culture and drug delivery. Recent reports have shown that amyloid fibril scaffolds generated from various proteins enhanced cell growth and proliferation of mammalian cells by interacting with the cell membrane and activating the cellular adhesion machinery.152,160,161 It also helps in the transportation of positively charged and hydrophobic therapeutic molecules such as methylene blue and riboflavin.162 Recently, aloe vera and bovine serum albumin (BSA) have been used for the development of a composite hydrogel. The developed composite hydrogel was tested in vitro and in vivo for its high efficiency in wound healing. Along with the development of the composite hydrogel, 3D printed conformation and bio-ink have been developed for wound dressing and tissue engineering applications.163,164 Also, macroscopic materials are being synthesized using amyloid fibrils, where the robust mechanical characteristics and chemical stability of amyloids have been harnessed to create macroscale materials such as hydrogels, macro-fibres, composites, sensors and matrices.152,165 The fibrillar structure of amyloid allows modification for the development of conductive materials that can be used as biodegradable and biocompatible conductive wires.166Amyloid fibrils can also be used as catalytic materials by introducing specific sites within the amyloid fibril structure that helps in providing a suitable microenvironment for catalytic reactions.167 Tyrosine and histidine side chains show esterase activity when they get a favourable microenvironment of self-assembled cross-β fibrils (HYHYHYHY). Histidine, owing to its potential for non-covalent interactions and hydrolase activity, plays a key role in esterase activity. Further, by modulating the activity through the tripeptide His–DPhe–DPhe, a thermally reversible hydrogel was formed with esterase activity. Also, an octapeptide-based hydrogel having hydrophobic activities of leucine and isoleucine has been reported for enhanced catalytic activities.165
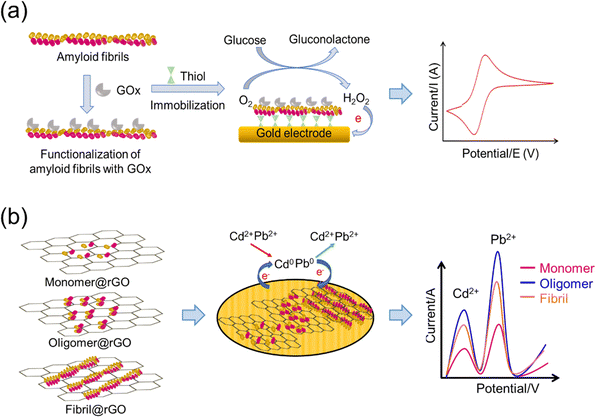
5.3. Amyloid-based materials in sensors
Self-assembled modified amyloid-based fibrils as nanomaterials have gained attention in various sensor applications.168–173 Amyloid-based sensors detect response due to alterations in amyloid conformation induced by the target analytes. These responses can be optical, electrochemical or mechanical and can be used for real-time detection and quantification of the target analytes (Fig. 4(a)). The application of biotin–streptavidin functionalized amyloids has been reported for the development of glucose sensors.174 Biotinylated whey protein-based amyloid fibrils have been prepared using succinimide-based biotin conjugates. The modified amyloids interacted with streptavidin-modified nanoparticles, quantum dots and glucose oxidase (GOx), where cyclic voltammetry was used to monitor glucose in the samples (Fig. 4(a)).175 The applications of amyloid fibrils in the development of colorimetric sensors has also been explored, where a colorimetric sensor was developed using an external stimuli-responsive fluorescence reporter molecule, 10,12-pentacosadiynoic acid, with α-synuclein amyloid fibrils.176 Also, an amyloid fibril-based colorimetric nanosensor has been reported for the detection of chromium(VI) using hen lysosome fibrils and acidified diphenylcarbazide as the colorimetric probe.177 The hydrogel of α-synuclein amyloid fibrils immobilized with horseradish peroxidase (HRP) enhanced the activity of immobilized HRP by four-fold, suggesting applications of enzyme–amyloid conjugates.178A gas sensor based on amyloid–reduced graphene oxide (rGO) nanocomposite materials has been developed and applied to e-textile yarns. The rGO/amyloid nanofibril/cotton yarns were used for the detection of NO2 gas owing to quick detection and high sensitivity and selectivity, which led to an e-textile wearable gas sensor.179 These studies suggested that amyloid nanofibrils can be used as molecular adhesives in the fabrication of sensitive sensors for personal healthcare or industrial purposes. A nanocomposite material based on an amyloid oligomer–reduced graphene oxide composite (AOrGOC) for developing an amperometric sensor has been reported to detect heavy metals using the stripping voltammetry technique (Fig. 4(b)).180,181 The sensor response of the amyloid fibrils generated from different proteins like lysozyme, bovine serum albumin and lactoglobulin has been investigated to determine the reactivity between amyloids and various heavy metal ions.
5.4. Nanofibrils as water purification materials
The problem of water contamination due to heavy metals is a global concern specifically for developing countries, necessitating the development of efficient and eco-friendly purification methods.182 The use of amyloid-based chelation for water purification has the potential to mitigate health risks associated with metal exposure and contribute to sustainable water treatment strategies.183 Due to distinct structural features, such as β-sheet rich conformations and exposed functional groups, amyloid fibrils have been reported for potential application in water purification.48,184–188 The ability of amyloids to sequester metals can be explored further for their potential as novel chelating agents and applications in environmental settings for the removal of metals.48,184 For effective and specific metal removal, the metal-binding properties of amyloid-derived peptides can be enhanced through modification of the amino acid sequences or introduction of specific functional groups. Hybrid materials along with amyloid-based chelators are being developed that can be incorporated into porous matrices or membranes for efficient removal of metal contaminants from water.189 For example, whey protein, a waste by-product from the cheese-processing industry, was used for preparing a functional scaffold for the β-lactoglobulin amyloid fibril/ZIF-8 (zeolitic imidazolate framework) based metal–organic framework for water purification and wastewater treatment.183,190 Owing to their hierarchical porous structure, the resultant aerogels based on amyloid fibrils with the ZIF-8 hybrid can successfully eliminate various types of heavy metals from water. Importantly, through adsorption–regeneration cycles, the hybrid aerogels can efficiently remove Hg2+ and Pb2+ from water.190 The reusable and sustainable materials from soy protein-based amyloids can also be used for manufacturing carbon aerogels having high removal efficacy for metals.191 The amyloid fibrils and their hybrids with modified properties can be a promising choice for efficient wastewater treatment.105,191Focused interventions to improve the elasticity, permeability and strength of amyloid fibril-based biomaterials by incorporating plasticizers, cross-linkers or polymers can further expand the field of their application.68 The major applications of amyloid-based biomaterials are in the fields of healthcare and sensing, where amyloid-based polymeric membranes are produced at an industrial scale for commercialization.152 It is essential to evaluate their stability and functionality to ensure effectiveness for safe use in patient-specific therapies by designing and incorporating specific drugs.195 Balanced production costs, optimal performance and safety considerations are critical to achieve sustainable and widely acceptable amyloid fibril-based biomaterials, as compared to conventionally used biomaterials.196 The emerging advances in the preparation and characterization of biocompatible and biodegradable amyloid fibril-based materials will open up new avenues for future applications of sustainable materials in healthcare and sensing.
Author contributions
Smriti Singh Yadav: writing of the original draft, conceptualization, visualization, writing – review & editing. Prabeen Kumar Padhy: visualization, writing – review & editing. Ashish Kumar Singh: visualization, writing – review & editing. Supriya Sharma: writing – review and editing. Tanu: writing – review & editing. Siraj Fatima: writing – review & editing. Anurag Sinha: writing – review & editing. Ramsha Tariq: writing – review & editing. Varsha: writing – review & editing. Sandeep Kumar Sharma: conceptualization, supervision, project administration, funding acquisition, writing – review & editing. Smriti Priya: conceptualization, supervision, project administration, funding acquisition, writing – review & editing.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors thank the Council of Scientific and Industrial Research, India for funding (HCP 31/4.1, CSIR India) and necessary facilities. SSY acknowledges the Senior Research Fellowship from ICMR-India. The CSIR-IITR manuscript communication number is IITR/SEC/MS/2023/86.References
- A. T. Nguyen, L. Parker, L. Brennan and S. Lockrey, J. Cleaner Prod., 2020, 252, 119792 CrossRef.
- I. Shahabi-Ghahfarrokhi, H. Almasi and A. Babaei-Ghazvini, Processing and development of polysaccharide-based biopolymers for packaging applications, Elsevier, 2020, pp.49–95 Search PubMed.
- L. Avérous and E. Pollet, Environmental silicate nano-biocomposites, Springer, 2012, pp.13–39 Search PubMed.
- J. F. Díaz-Villanueva, R. Díaz-Molina and V. García-González, Int. J. Mol. Sci., 2015, 16, 17193–17230 CrossRef PubMed.
- S. M. Choi, P. Chaudhry, S. M. Zo and S. S. Han, Cutting-edge enabling technologies for regenerative medicine, 2018, pp.161–210 Search PubMed.
- A. Tuwalska, S. Grabska-Zielińska and A. Sionkowska, Polymers, 2022, 14, 1343 CrossRef CAS PubMed.
- J. Majorošová, N. Tomašovičová, V. Gdovinová, C.-W. Yang, M. Batkova, I. Batko, M. Demčaková, K. Csach, M. Kubovčíková and S. Hayryan, J. Magn. Magn. Mater., 2019, 471, 400–405 CrossRef.
- S. Devi, M. Chaturvedi, S. Fatima and S. Priya, Toxicology, 2022, 465, 153049 CrossRef CAS PubMed.
- N. Cremades and C. M. Dobson, Neurobiol. Dis., 2018, 109, 178–190 CrossRef CAS PubMed.
- U. Sengupta and R. Kayed, Prog. Neurobiol., 2022, 214, 102270 CrossRef CAS PubMed.
- P. Taboada, S. Barbosa, J. Juárez, M. A. Meda and V. Mosquera, Proteins in Solution and at Interfaces: Methods and Applications in Biotechnology and Materials Science, 2013, pp.233–282 Search PubMed.
- K. J. Y. Low, A. Venkatraman, J. S. Mehta and K. Pervushin, J. Adv. Res., 2022, 36, 113–132 CrossRef CAS PubMed.
- J. D. Sipe, M. D. Benson, J. N. Buxbaum, S.-I. Ikeda, G. Merlini, M. J. Saraiva and P. Westermark, Amyloid, 2016, 23, 209–213 CrossRef CAS PubMed.
- N. Louros, J. Schymkowitz and F. Rousseau, Nat. Rev. Mol. Cell Biol., 2023, 24, 912–933 CrossRef CAS PubMed.
- F. Chiti and C. M. Dobson, Annu. Rev. Biochem., 2017, 86, 27–68 CrossRef CAS PubMed.
- Y. Cao and R. Mezzenga, Adv. Colloid Interface Sci., 2019, 269, 334–356 CrossRef CAS PubMed.
- K. Antonets and A. Nizhnikov, Prion, 2017, 11, 300–312 CrossRef CAS PubMed.
- P. Seth, A. Mukherjee and N. Sarkar, Int. J. Biol. Macromol., 2023, 253, 127177 CrossRef CAS PubMed.
- J. P. Leite, F. Figueira, R. F. Mendes, F. A. Almeida Paz and L. Gales, ACS Sens., 2023, 8, 1033–1053 CrossRef CAS PubMed.
- Y.-R. Lai, S. S.-S. Wang, T.-L. Hsu, S.-H. Chou, S.-C. How and T.-H. Lin, Polymers, 2023, 15, 1444 CrossRef CAS PubMed.
- C. A. Hauser, S. Maurer-Stroh and I. C. Martins, Chem. Soc. Rev., 2014, 43, 5326–5345 RSC.
- J. Zhao and P. Yang, Adv. Mater. Interfaces, 2020, 7, 2001060 CrossRef CAS.
- H. Chen, J. Wang, Y. Cheng, C. Wang, H. Liu, H. Bian, Y. Pan, J. Sun and W. Han, Polymers, 2019, 11, 2039 CrossRef CAS PubMed.
- L. Liu, S. Yang, C. Chen, Y. Fang, L. Li and Z. Ban, Food Packag. Shelf Life, 2023, 37, 101080 CrossRef CAS.
- S. Kim, J. H. Kim, J. S. Lee and C. B. Park, Small, 2015, 11, 3623–3640 CrossRef CAS PubMed.
- Z. L. Almeida and R. M. Brito, Molecules, 2020, 25, 1195 CrossRef CAS PubMed.
- L. Breydo and V. N. Uversky, Bio-nanoimaging, Elsevier, 2014, pp. 1–14 DOI:10.1016/b978-0-12-394431-3.00001-8.
- E. Chatani, K. Yuzu, Y. Ohhashi and Y. Goto, Int. J. Mol. Sci., 2021, 22, 4349 CrossRef CAS PubMed.
- G. Wei, Z. Su, N. P. Reynolds, P. Arosio, I. W. Hamley, E. Gazit and R. Mezzenga, Chem. Soc. Rev., 2017, 46, 4661–4708 RSC.
- B. H. Toyama and J. S. Weissman, Annu. Rev. Biochem., 2011, 80, 557–585 CrossRef CAS PubMed.
- J. S. Pedersen and D. E. Otzen, Protein Sci., 2008, 17, 2–10 CrossRef CAS PubMed.
- J. Park, B. Kahng and W. Hwang, PLoS Comput. Biol., 2009, 5, e1000492 CrossRef PubMed.
- S. Y. Ow and D. E. Dunstan, Protein Sci., 2014, 23, 1315–1331 CrossRef CAS PubMed.
- T. R. Jahn, O. S. Makin, K. L. Morris, K. E. Marshall, P. Tian, P. Sikorski and L. C. Serpell, J. Mol. Biol., 2010, 395, 717–727 CrossRef CAS PubMed.
- K. Gade Malmos, L. M. Blancas-Mejia, B. Weber, J. Buchner, M. Ramirez-Alvarado, H. Naiki and D. Otzen, Amyloid, 2017, 24, 1–16 CrossRef CAS PubMed.
- R. Jurado Palomares and N. Gálvez Rodríguez, 2021.
- J. R. Engen and E. A. Komives, Trends Biochem. Sci., 2020, 45, 906–918 CrossRef CAS PubMed.
- H. Zhang, L.-Q. Xu and S. Perrett, Methods, 2011, 53, 285–294 CrossRef CAS PubMed.
- C. Lendel and N. Solin, RSC Adv., 2021, 11, 39188–39215 RSC.
- T. Srivastava, R. Raj, A. Dubey, D. Kumar, R. K. Chaturvedi, S. K. Sharma and S. Priya, Sci. Rep., 2020, 10, 18412 CrossRef CAS PubMed.
- Y. Cao, J. Adamcik, M. Diener, J. R. Kumita and R. Mezzenga, J. Am. Chem. Soc., 2021, 143, 11473–11481 CrossRef CAS PubMed.
- N. J. Greenfield, Nat. Protoc., 2006, 1, 2876–2890 CrossRef CAS PubMed.
- O. Ozohanics and A. Ambrus, Life, 2020, 10(11), 286–305 CrossRef CAS PubMed.
- B. Li, P. Ge, K. A. Murray, P. Sheth, M. Zhang, G. Nair, M. R. Sawaya, W. S. Shin, D. R. Boyer and S. Ye, Nat. Commun., 2018, 9, 3609 CrossRef PubMed.
- W. Zhang, A. Tarutani, K. L. Newell, A. G. Murzin, T. Matsubara, B. Falcon, R. Vidal, H. J. Garringer, Y. Shi and T. Ikeuchi, Nature, 2020, 580, 283–287 CrossRef CAS PubMed.
- F. Wahid, X.-J. Zhao, S.-R. Jia, H. Bai and C. Zhong, Composites, Part B, 2020, 200, 108208 CrossRef CAS.
- Y. Shen, A. Levin, A. Kamada, Z. Toprakcioglu, M. Rodriguez-Garcia, Y. Xu and T. P. Knowles, ACS Nano, 2021, 15, 5819–5837 CrossRef CAS PubMed.
- S. Bolisetty and R. Mezzenga, Nat. Nanotechnol., 2016, 11, 365–371 CrossRef CAS PubMed.
- Q. Zhang, S. Bolisetty, Y. Cao, S. Handschin, J. Adamcik, Q. Peng and R. Mezzenga, Angew. Chem., Int. Ed., 2019, 58, 6012–6016 CrossRef CAS PubMed.
- A. C. Lin, F. Xie, R. Chang, N. Beaver, C. Drewery, C. Collins, C. Lehr, E. M. Jones and S. Zhang, Carbon Trends, 2021, 5, 100135 CrossRef CAS.
- T. Li, J. Zhou, M. Peydayesh, Y. Yao, M. Bagnani, I. Kutzli, Z. Chen, L. Wang and R. Mezzenga, Adv. Sustainable Syst., 2023, 7, 2200414 CrossRef CAS.
- Y. A. Shah, S. Bhatia, A. Al-Harrasi, M. Afzaal, F. Saeed, M. K. Anwer, M. R. Khan, M. Jawad, N. Akram and Z. Faisal, Polymers, 2023, 15(7), 1724 CrossRef CAS PubMed.
- S. Sid, R. S. Mor, A. Kishore and V. S. Sharanagat, Trends Food Sci. Technol., 2021, 115, 87–104 CrossRef CAS.
- A. R. Ferreira, V. D. Alves and I. M. Coelhoso, Membranes, 2016, 6, 22 CrossRef PubMed.
- R. Stefani, G. L. Vinhal, D. V. do Nascimento, M. C. S. Pereira, P. B. Pertuzatti and K. da Silva Chaves, Industrial Applications for Intelligent Polymers and Coatings, 2016, pp.253–269 Search PubMed.
- N. K. Dubey and R. Dubey, Biopolymer-based formulations, Elsevier, 2020, pp.675–695 Search PubMed.
- S. A. Mohamed, M. El-Sakhawy and M. A.-M. El-Sakhawy, Carbohydr. Polym., 2020, 238, 116178 CrossRef CAS PubMed.
- A. E. Quirós-Sauceda, J. F. Ayala-Zavala, G. I. Olivas and G. A. González-Aguilar, J. Food Sci. Technol., 2014, 51, 1674–1685 CrossRef PubMed.
- V. M. Azevedo, S. V. Borges, J. M. Marconcini, M. I. Yoshida, A. R. S. Neto, T. C. Pereira and C. F. G. Pereira, Carbohydr. Polym., 2017, 157, 971–980 CrossRef CAS PubMed.
- B. E. Teixeira-Costa and C. T. Andrade, Polysaccharides, 2022, 3, 32–58 CrossRef CAS.
- I. Assad, S. U. Bhat, A. Gani and A. Shah, Int. J. Biol. Macromol., 2020, 164, 707–716 CrossRef CAS PubMed.
- S. Y. J. Sim, A. Srv, J. H. Chiang and C. J. Henry, Foods, 2021, 10, 1967 CrossRef CAS PubMed.
- T. Jiang, Q. Duan, J. Zhu, H. Liu and L. Yu, Adv. Ind. Eng. Polym. Res., 2020, 3, 8–18 Search PubMed.
- H. Abdul Khalil, A. Banerjee, C. K. Saurabh, Y. Tye, A. Suriani, A. Mohamed, A. Karim, S. Rizal and M. Paridah, Food Eng. Rev., 2018, 10, 139–153 CrossRef CAS.
- U. Amin, M. U. Khan, Y. Majeed, M. Rebezov, M. Khayrullin, E. Bobkova, M. A. Shariati, I. M. Chung and M. Thiruvengadam, Int. J. Biol. Macromol., 2021, 183, 2184–2198 CrossRef CAS PubMed.
- L. P. Datta, S. Manchineella and T. Govindaraju, Biomaterials, 2020, 230, 119633 CrossRef CAS PubMed.
- M. Kumar, M. Tomar, S. Punia, J. Dhakane-Lad, S. Dhumal, S. Changan, M. Senapathy, M. K. Berwal, V. Sampathrajan and A. A. Sayed, LWT–Food Sci. Technol., 2022, 154, 112620 CrossRef CAS.
- S. J. Calva-Estrada, M. Jiménez-Fernández and E. Lugo-Cervantes, Food Eng. Rev., 2019, 11, 78–92 CrossRef CAS.
- M. G. A. Vieira, M. A. Da Silva, L. O. Dos Santos and M. M. Beppu, Eur. Polym. J., 2011, 47, 254–263 CrossRef CAS.
- K. Dangaran, P. M. Tomasula and P. Qi, Edible films and coatings for food applications, 2009, pp.25–56 Search PubMed.
- M. Wihodo and C. I. Moraru, J. Food Eng., 2013, 114, 292–302 CrossRef CAS.
- A. Samir, F. H. Ashour, A. A. Hakim and M. Bassyouni, npj Mater. Degrad., 2022, 6, 68 CrossRef CAS.
- S. Khansari, S. Sinha-Ray, A. L. Yarin and B. Pourdeyhimi, Ind. Eng. Chem. Res., 2013, 52, 15104–15113 CrossRef CAS.
- D. Chen, O. G. Jones and O. H. Campanella, Crit. Rev. Food Sci. Nutr., 2023, 63, 4554–4578 CrossRef CAS PubMed.
- M. Hadidi, S. Jafarzadeh, M. Forough, F. Garavand, S. Alizadeh, A. Salehabadi, A. M. Khaneghah and S. M. Jafari, Trends Food Sci. Technol., 2022, 120, 154–173 CrossRef CAS.
- S. Tortorella, M. Maturi, V. V. Buratti, G. Vozzolo, E. Locatelli, L. Sambri and M. C. Franchini, RSC Adv., 2021, 11, 39004–39026 RSC.
- J. Gómez-Estaca, R. Gavara, R. Catala and P. Hernández-Muñoz, Packag. Technol. Sci., 2016, 29, 203–224 CrossRef.
- S. Jafarzadeh, M. Forough, S. Amjadi, V. Javan Kouzegaran, H. Almasi, F. Garavand and M. Zargar, Crit. Rev. Food Sci. Nutr., 2023, 63, 9667–9693 CrossRef CAS PubMed.
- A. R. Hammam, SN Appl. Sci., 2019, 1, 1–11 Search PubMed.
- R. Bhaskar, S. M. Zo, B. N. Kanan, S. Purohit, M. K. Gupta and S. S. Han, Polym. Test., 2023, 108097 CrossRef CAS.
- J.-M. Fan, W. Ma, G.-Q. Liu, S.-W. Yin, C.-H. Tang and X.-Q. Yang, Food Hydrocolloids, 2014, 36, 60–69 CrossRef CAS.
- M. Moghadam, M. Salami, M. Mohammadian, M. Khodadadi and Z. Emam-Djomeh, Food Hydrocolloids, 2020, 104, 105735 CrossRef CAS.
- S. Rojas-Lema, K. Nilsson, M. Langton, J. Trifol, J. Gomez-Caturla, R. Balart, D. Garcia-Garcia and R. Moriana, J. Food Eng., 2023, 339, 111282 CrossRef CAS.
- T. Zheng, X. Yu and S. Pilla, Carbohydr. Polym., 2017, 157, 1333–1340 CrossRef CAS PubMed.
- D. Nataraj, P. Aramwit, G. Nagananda and N. Reddy, Eur. Polym. J., 2020, 134, 109800 CrossRef CAS.
- N. Kumari, S. P. Bangar, M. Petrů, R. Ilyas, A. Singh and P. Kumar, Foods, 2021, 10, 1976 CrossRef CAS PubMed.
- C. H. Tang, M. L. Xiao, Z. Chen and X. Q. Yang, J. Appl. Polym. Sci., 2011, 122, 789–797 CrossRef CAS.
- T. Zhong, Y. Liang, S. Jiang, L. Yang, Y. Shi, S. Guo and C. Zhang, RSC Adv., 2017, 7, 41610–41618 RSC.
- L. Sharma, H. K. Sharma and C. S. Saini, J. Food Sci. Technol., 2018, 55, 532–539 CrossRef CAS PubMed.
- A. Osorio-Ruiz, R. J. Avena-Bustillos, B.-S. Chiou, F. Rodríguez-González and A.-L. Martinez-Ayala, ACS Omega, 2019, 4, 19172–19176 CrossRef CAS PubMed.
- M.-N. Efthymiou, E. Tsouko, A. Papagiannopoulos, I.-G. Athanasoulia, M. Georgiadou, S. Pispas, D. Briassoulis, T. Tsironi and A. Koutinas, Sci. Rep., 2022, 12, 6935 CrossRef CAS PubMed.
- K. K. Dash, A. Kumar, S. Kumari and M. A. Malik, J. Polym. Environ., 2021, 29, 3649–3659 CrossRef CAS.
- C. Zhang, Z. Wang, Y. Li, Y. Yang, X. Ju and R. He, Food Chem., 2019, 272, 694–701 CrossRef CAS PubMed.
- T. G. d Oliveira, G. L. d A. Makishi, H. Chambi, A. M. Q. B. Bittante, R. V. Lourenço and P. J. d A. Sobral, Ind. Crops Prod., 2015, 67, 355–363 CrossRef CAS.
- S. G. Giteru, I. Oey, M. A. Ali, S. K. Johnson and Z. Fang, Food Control, 2017, 80, 37–44 CrossRef CAS.
- M. Dong, L. Tian, J. Li, J. Jia, Y. Dong, Y. Tu, X. Liu, C. Tan and X. Duan, LWT–Food Sci. Technol, 2022, 154, 112868 CrossRef CAS.
- M. Habibi Zarabadi, M. Kadivar and J. Keramat, J. Food Process. Preserv., 2018, 42, e13513 CrossRef.
- Q. Yang, A. Lue, H. Qi, Y. Sun, X. Zhang and L. Zhang, Macromol. Biosci., 2009, 9, 849–856 CrossRef CAS PubMed.
- H. Xie, Y. Wang, K. Ouyang, L. Zhang, J. Hu, S. Huang, W. Sun, P. Zhang, H. Xiong and Q. Zhao, Int. J. Biol. Macromol., 2023, 236, 123877 CrossRef CAS PubMed.
- H. A. Razzaq, M. Pezzuto, G. Santagata, C. Silvestre, S. Cimmino, N. Larsen and D. Duraccio, Food Hydrocolloids, 2016, 58, 276–283 CrossRef CAS.
- N. A. Mir, C. S. Riar and S. Singh, Food Hydrocolloids, 2023, 135, 108190 CrossRef CAS.
- M. C. Condés, M. C. Añón, A. N. Mauri and A. Dufresne, Food Hydrocolloids, 2015, 47, 146–157 CrossRef.
- R. Thakur, R. Santhosh, Y. Kumar, V. R. Suryavanshi, H. Singhi, D. Madhubabu, S. Wickramarachchi, K. Pal and P. Sarkar, Trends Food Sci. Technol., 2023, 104143 CrossRef CAS.
- S. Rahman, J. Gogoi, S. Dubey and D. Chowdhury, Int. J. Biol. Macromol., 2023, 128197 Search PubMed.
- Z. A. Maryam Adilah and Z. A. Nur Hanani, Food Biosci., 2016, 16, 66–71 CrossRef CAS.
- F. Liu, R. J. Avena-Bustillos, B.-S. Chiou, Y. Li, Y. Ma, T. G. Williams, D. F. Wood, T. H. McHugh and F. Zhong, Food Hydrocolloids, 2017, 62, 212–221 CrossRef CAS.
- M. Jridi, O. Abdelhedi, N. Zouari, N. Fakhfakh and M. Nasri, Food Hydrocolloids, 2019, 89, 370–378 CrossRef CAS.
- Y. Lu, Q. Luo, Y. Chu, N. Tao, S. Deng, L. Wang and L. Li, Polymers, 2022, 14, 436 CrossRef CAS PubMed.
- T. Zheng, P. Tang and G. Li, Int. J. Biol. Macromol., 2023, 241, 124494 CrossRef CAS PubMed.
- P. Tang, T. Zheng, C. Yang and G. Li, Food Chem., 2022, 393, 133353 CrossRef CAS PubMed.
- S. Nuanmano, T. Prodpran and S. Benjakul, Food Hydrocolloids, 2015, 47, 61–68 CrossRef CAS.
- L. Bonnaillie, H. Zhang, S. Akkurt, K. Yam and P. Tomasula, Polymers, 2014, 6, 2018–2036 CrossRef.
- S. Bhatia, A. Al-Harrasi, M. S. Al-Azri, S. Ullah, H. A. Makeen, A. M. Meraya, M. Albratty, A. Najmi and M. K. Anwer, Polymers, 2022, 14, 4065 CrossRef CAS PubMed.
- M. d R. Moreira, M. Pereda, N. E. Marcovich and S. I. Roura, J. Food Sci., 2011, 76, M54–M63 CAS.
- S. Kandasamy, J. Yoo, J. Yun, H.-B. Kang, K.-H. Seol, H.-W. Kim and J.-S. Ham, Coatings, 2021, 11, 1056 CrossRef CAS.
- A. S. Silva, E. C. Costa, S. Reis, C. Spencer, R. C. Calhelha, S. P. Miguel, M. P. Ribeiro, L. Barros, J. A. Vaz and P. Coutinho, Polymers, 2022, 14, 4931 CrossRef CAS PubMed.
- R. Pandey, S. Dixit and R. Dubey, Novel Sustainable Raw Material Alternatives for the Textiles and Fashion Industry, Springer, 2023, pp.57–79 Search PubMed.
- R. I. Kunz, R. M. C. Brancalhão, L. de Fátima Chasko Ribeiro and M. R. M. Natali, BioMed Res. Int., 2016, 2016, 8175701 Search PubMed.
- S.-J. Seo, G. Das, H.-S. Shin and J. K. Patra, Int. J. Mol. Sci., 2023, 24, 4951 CrossRef CAS PubMed.
- L. Fuentes-Mera, Res. Dev. Mater. Sci., 2019, 10, 1–10 Search PubMed.
- N. Kono, H. Nakamura, A. Tateishi, K. Numata and K. Arakawa, Zool. Lett., 2021, 7, 1–9 CrossRef PubMed.
- M. H. Khosropanah, M. A. Vaghasloo, M. Shakibaei, A. L. Mueller, A. M. Kajbafzadeh, L. Amani, I. Haririan, A. Azimzadeh, Z. Hassannejad and M. M. Zolbin, J. Tissue Eng. Regener. Med., 2022, 16, 91–109 CrossRef CAS PubMed.
- A. Sammi, Divya, S. Mahapatra, R. Kumar and P. Chandra, Biotechnol. Bioeng., 2022, 119, 784–806 CrossRef CAS PubMed.
- P. Wongpanit, O. Pornsunthorntawee and R. Rujiravanit, Nat. Polym., 2012, 219–222 CAS.
- J. K. Sahoo, O. Hasturk, T. Falcucci and D. L. Kaplan, Nat. Rev. Chem., 2023, 1–17 Search PubMed.
- B. Joseph and S. J. Raj, Front. Life Sci., 2012, 6, 55–60 CrossRef CAS.
- R. Fedic, M. Žurovec and F. E. Sehnal, J. Biol. Chem., 2003, 278, 35255–35264 CrossRef CAS PubMed.
- D. Wilson, R. Valluzzi and D. Kaplan, Biophys. J., 2000, 78, 2690–2701 CrossRef CAS PubMed.
- R. Fedič, M. Žurovec and F. Sehnal, J. Biol. Chem., 2003, 278, 35255–35264 CrossRef PubMed.
- W. Sun, D. A. Gregory, M. A. Tomeh and X. Zhao, Int. J. Mol. Sci., 2021, 22, 1499 CrossRef CAS PubMed.
- S. Grabska-Zielińska and A. Sionkowska, Materials, 2021, 14, 1510 CrossRef PubMed.
- C. Zhang, Y. Zhang, H. Shao and X. Hu, ACS Appl. Mater. Interfaces, 2016, 8, 3349–3358 CrossRef CAS PubMed.
- W. Song, M. Muthana, J. Mukherjee, R. J. Falconer, C. A. Biggs and X. Zhao, ACS Biomater. Sci. Eng., 2017, 3, 1027–1038 CrossRef CAS PubMed.
- W. Sun, Y. Zhang, D. A. Gregory, A. Jimenez-Franco, M. A. Tomeh, S. Lv, J. Wang, J. W. Haycock, J. R. Lu and X. Zhao, Prog. Nat. Sci., 2020, 30, 686–696 CrossRef CAS.
- J. Melke, S. Midha, S. Ghosh, K. Ito and S. Hofmann, Acta Biomater., 2016, 31, 1–16 CrossRef CAS PubMed.
- M. Saric and T. Scheibel, Curr. Opin. Biotechnol, 2019, 60, 213–220 CrossRef CAS PubMed.
- B. Marelli, M. Brenckle, D. L. Kaplan and F. G. Omenetto, Sci. Rep., 2016, 6, 25263 CrossRef CAS PubMed.
- F. V. Dos Santos, R. L. Siqueira, L. de Morais Ramos, S. A. Yoshioka, M. C. Branciforti and D. S. Correa, Int. J. Biol. Macromol., 2024, 254, 127641 CrossRef CAS PubMed.
- J. Liu, L. Shi, Y. Deng, M. Zou, B. Cai, Y. Song, Z. Wang and L. Wang, Biomaterials, 2022, 121638 CrossRef CAS PubMed.
- W. Li, Z. Huang, R. Cai, W. Yang, H. He and Y. Wang, Int. J. Mol. Sci., 2020, 22, 105 CrossRef CAS PubMed.
- R. Wang, J. Li, W. Chen, T. Xu, S. Yun, Z. Xu, Z. Xu, T. Sato, B. Chi and H. Xu, Adv. Funct. Mater., 2017, 27, 1604894 CrossRef.
- G. Tao, R. Cai, Y. Wang, L. Liu, H. Zuo, P. Zhao, A. Umar, C. Mao, Q. Xia and H. He, Mater. Des., 2019, 180, 107940 CrossRef CAS.
- M. K. DeBari, M. N. Keyser, M. A. Bai and R. D. Abbott, Connect. Tissue Res., 2020, 61, 163–173 CrossRef CAS PubMed.
- S. M. Yukseloglu, N. Sokmen and S. Canoglu, Microelectron. Eng., 2015, 146, 43–47 CrossRef CAS.
- C. Belda Marín, V. Fitzpatrick, D. L. Kaplan, J. Landoulsi, E. Guénin and C. Egles, Front. Chem., 2020, 8, 604398 CrossRef PubMed.
- G. Tao, R. Cai, Y. Wang, H. Zuo and H. He, Mater. Sci. Eng., C, 2021, 119, 111597 CrossRef CAS PubMed.
- M. Yang, Y. Wang, G. Tao, R. Cai, P. Wang, L. Liu, L. Ai, H. Zuo, P. Zhao, A. Umar, C. Mao and H. He, Nanomaterials, 2018, 8(4), 235 CrossRef PubMed.
- Z. Wang, Y. Zhang, J. Zhang, L. Huang, J. Liu, Y. Li, G. Zhang, S. C. Kundu and L. Wang, Sci. Rep., 2014, 4, 7064 CrossRef PubMed.
- Y. Song, C. Zhang, J. Zhang, N. Sun, K. Huang, H. Li, Z. Wang, K. Huang and L. Wang, Acta Biomater., 2016, 41, 210–223 CrossRef CAS PubMed.
- Z. Wang, J. Wang, Y. Jin, Z. Luo, W. Yang, H. Xie, K. Huang and L. Wang, ACS Appl. Mater. Interfaces, 2015, 7, 24629–24640 CrossRef CAS PubMed.
- E. Arslan, I. C. Garip, G. Gulseren, A. B. Tekinay and M. O. Guler, Adv. Healthcare Mater., 2014, 3, 1357–1376 CrossRef CAS PubMed.
- S. Chowdhury and N. Sarkar, Biotechnol. Bioeng., 2024, 121, 26–38 CrossRef CAS PubMed.
- M. Lassé, D. Ulluwishewa, J. Healy, D. Thompson, A. Miller, N. Roy, K. Chitcholtan and J. A. Gerrard, Food Chem., 2016, 192, 491–498 CrossRef PubMed.
- F. D. Victorelli, C. F. Rodero, V. Lutz-Bueno, M. Chorilli and R. Mezzenga, Adv. Healthcare Mater., 2023, 12, 2202720 CrossRef CAS PubMed.
- J. Boekhoven, M. Koot, T. A. Wezendonk, R. Eelkema and J. H. van Esch, J. Am. Chem. Soc., 2012, 134, 12908–12911 CrossRef CAS PubMed.
- T. Wu, Q. Jiang, D. Wu, Y. Hu, S. Chen, T. Ding, X. Ye, D. Liu and J. Chen, Food Chem., 2019, 274, 698–709 CrossRef CAS PubMed.
- U. Shimanovich, I. Efimov, T. O. Mason, P. Flagmeier, A. K. Buell, A. Gedanken, S. Linse, K. S. Åkerfeldt, C. M. Dobson and D. A. Weitz, ACS Nano, 2015, 9, 43–51 CrossRef CAS PubMed.
- A. Kumari and B. Ahmad, RSC Adv., 2019, 9, 37424–37435 RSC.
- L. Yang, H. Li, L. Yao, Y. Yu and G. Ma, ACS Omega, 2019, 4, 8071–8080 CrossRef CAS PubMed.
- S. Chowdhury and N. Sarkar, Biotechnol. Bioeng., 2024, 121, 26–38 CrossRef CAS PubMed.
- R. S. Jacob, S. Das, N. Singh, K. Patel, D. Datta, S. Sen and S. K. Maji, Biochemical and Biophysical Roles of Cell Surface Molecules, 2018, pp.79–97 Search PubMed.
- Y. R. Lai, S. S. Wang, T. L. Hsu, S. H. Chou, S. C. How and T. H. Lin, Polymers, 2023, 15, 1444 CrossRef CAS PubMed.
- K. Naik, P. Singh, M. Yadav, S. K. Srivastava, S. Tripathi, R. Ranjan, P. Dhar, A. K. Verma, S. Chaudhary and A. S. Parmar, J. Mater. Chem. B, 2023, 11, 8142–8158 RSC.
- H. Taneja, S. M. Salodkar, A. Singh Parmar and S. Chaudhary, J. Mol. Liq., 2022, 367, 120390 CrossRef CAS.
- J. Li and F. Zhang, Int. J. Mol. Sci., 2021, 22, 10698 CrossRef CAS PubMed.
- J. Li and F. Zhang, Int. J. Mol. Sci., 2021, 22, 10698 CrossRef CAS PubMed.
- C. M. Rufo, Y. S. Moroz, O. V. Moroz, J. Stöhr, T. A. Smith, X. Hu, W. F. DeGrado and I. V. Korendovych, Nat. Chem., 2014, 6, 303–309 CrossRef CAS PubMed.
- G. Wei, Z. Su, N. P. Reynolds, P. Arosio, I. W. Hamley, E. Gazit and R. Mezzenga, Chem. Soc. Rev., 2017, 46, 4661–4708 RSC.
- T. Li, X.-M. Lu, M.-R. Zhang, K. Hu and Z. Li, Bioact. Mater., 2022, 11, 268–282 CAS.
- Y. Wu, F. Wang, K. Lu, M. Lv and Y. Zhao, Sens. Actuators, B, 2017, 244, 1022–1030 CrossRef CAS.
- J. Z. Hassan, A. Raza, Z. U. Din Babar, U. Qumar, N. T. Kaner and A. Cassinese, J. Mater. Chem. A, 2023, 11, 6016–6063 RSC.
- C. A. E. Hauser, S. Maurer-Stroh and I. C. Martins, Chem. Soc. Rev., 2014, 43, 5326 RSC.
- S. Mankar, A. Anoop, S. Sen and S. K. Maji, Nano Rev., 2011, 2, 6032 CrossRef PubMed.
- M. Díaz-Caballero, S. Navarro and S. Ventura, Biomacromolecules, 2021, 22, 2822–2833 CrossRef PubMed.
- L. Sasso, S. Suei, L. Domigan, J. Healy, V. Nock, M. A. K. Williams and J. A. Gerrard, Nanoscale, 2014, 6, 1629–1634 RSC.
- J. E. Yang, J. S. Park, E. Cho, S. Jung and S. R. Paik, Langmuir, 2015, 31, 1802–1810 CrossRef CAS PubMed.
- W.-H. Leung, L. Zou, W.-H. Lo and P.-H. Chan, ChemPlusChem, 2013, 78, 1440 CrossRef CAS PubMed.
- G. Bhak, S. Lee, J. W. Park, S. Cho and S. R. Paik, Biomaterials, 2010, 31, 5986–5995 CrossRef CAS PubMed.
- S. W. Lee, W. Lee, I. Kim, D. Lee, D. Park, W. Kim, J. Park, J. H. Lee, G. Lee and D. S. Yoon, ACS Sens., 2021, 6, 777–785 CrossRef CAS PubMed.
- W. Li, R. Chen, W. Qi, L. Cai, Y. Sun, M. Sun, C. Li, X. Yang, L. Xiang, D. Xie and T. Ren, ACS Sens., 2019, 4, 2809–2818 CrossRef CAS PubMed.
- C. Kim, J. Park, W. Kim, W. Lee, S. Na and J. Park, Bioelectrochemistry, 2022, 147, 108214 CrossRef CAS PubMed.
- K. H. Vardhan, P. S. Kumar and R. C. Panda, J. Mol. Liq., 2019, 290, 111197 CrossRef CAS.
- M. Peydayesh, X. Chen, J. Vogt, F. Donat, C. R. Müller and R. Mezzenga, Chem. Commun., 2022, 58, 5104–5107 RSC.
- M. Peydayesh and R. Mezzenga, Nat. Commun., 2021, 12, 3248 CrossRef CAS PubMed.
- S. Bolisetty, N. Reinhold, C. Zeder, M. N. Orozco and R. Mezzenga, Chem. Commun., 2017, 53, 5714–5717 RSC.
- X. Zhang, M. R. Razanajatovo, X. Du, S. Wang, L. Feng, S. Wan, N. Chen and Q. Zhang, Eco-Environ. Health, 2023, 264–277 CrossRef CAS PubMed.
- W. L. Soon, M. Peydayesh, R. Mezzenga and A. Miserez, Chem. Eng. J., 2022, 445, 136513 CrossRef CAS.
- X. Jia, M. Peydayesh, Q. Huang and R. Mezzenga, Small, 2022, 18, 2105502 CrossRef CAS PubMed.
- C. Liang, L. Zhao, L. Qiao and K. Du, J. Hazard. Mater., 2022, 425, 127886 CrossRef CAS PubMed.
- X. Jia, M. Peydayesh, Q. Huang and R. Mezzenga, Small, 2022, 18, 2105502 CrossRef CAS PubMed.
- M. Peydayesh, J. Vogt, X. Chen, J. Zhou, F. Donat, M. Bagnani, C. R. Müller and R. Mezzenga, Chem. Eng. J., 2022, 449, 137703 CrossRef CAS.
- T. Oz, A. Kaushik and M. Kujawska, Mater. Adv., 2023, 6464–6477 RSC.
- Q. Zheng, J. Yao, Z. Sun, Y. Mao, J. Wei, Y. Xie, X. K. K. Hu and X. Li, Mater. Adv., 2024 10.1039/D3MA01046E.
- Y. Ni, Y. Jiang, K. Wang, Z. Shao, X. Chen, S. Sun, H. Yu and W. Li, Int. J. Artif. Organs, 2019, 42, 31–41 CrossRef CAS PubMed.
- G. Kaur, G. Narayanan, D. Garg, A. Sachdev and I. Matai, ACS Appl. Bio Mater., 2022, 5, 2069–2106 CrossRef CAS PubMed.
- E. Abaie, L. Xu and Y.-X. Shen, Front. Environ. Sci. Eng., 2021, 15, 1–33 CrossRef.
| This journal is © The Royal Society of Chemistry 2024 |