Gallium-based metal–organic frameworks loaded with antimicrobial peptides for synergistic killing of drug-resistant bacteria†
Shuo
Liu
abc,
Yuxin
Ji
a,
Hangqi
Zhu
a,
Zhishang
Shi
 a,
Mingchun
Li
a and
Qilin
Yu
a,
Mingchun
Li
a and
Qilin
Yu
 *ac
*ac
aNational Key Laboratory of Intelligent Tracking and Forecasting for Infectious Diseases, College of Life Sciences, Nankai University, Tianjin 300071, China
bCollege of Environmental Science and Engineering, Tianjin Key Laboratory of Environmental Remediation and Pollution Control, Nankai University, Tianjin 300350, China
cResearch Center for Infectious Diseases, Nankai University, Tianjin 300350, China. E-mail: yuqilin@mail.nankai.edu.cn
First published on 12th October 2023
Abstract
Increased antibiotic resistance has made bacterial infections a global concern, which requires novel non-antibiotic-dependent antibacterial strategies to address the menace. Antimicrobial peptides (AMPs) are a promising antibiotic alternative, whose antibacterial mechanism is mainly to destroy the membrane of bacteria. Gallium ions exhibit an antibacterial effect by interfering with the iron metabolism of bacteria. With the rapid development of nanotechnology, it is worth studying the potential of gallium-AMP-based nanocomposites for treating bacterial infections. Herein, novel gallium-based metal–organic frameworks (MOFs) were synthesized at room temperature, followed by in situ loading of the model AMP melittin. The obtained nanocomposites exhibited stronger antibacterial activity than pure MEL and gallium ions, achieving the effects of “one plus one is greater than two”. Moreover, the nanocomposites showed favorable biocompatibility and accelerated healing of a wound infected by methicillin-resistant Staphylococcus aureus by down-regulation of inflammatory cytokines IL-6 and TNF-α. This work presents an innovative antibacterial strategy to overcome the antibiotic resistance crisis and expand the application of AMPs.
1. Introduction
Bacterial infections have become one of the top ten causes of death, posing a serious threat to human health.1 As the first line of immune defence for the human body, healthy skin can prevent bacteria and other pathogenic microorganisms from invading the body. After skin injury, bacteria can easily invade the body through the wound, so, it is important to treat the wound in a timely fashion and correctly.2–4 Antibiotics are the preferred method for treating bacterial infections, but improper use of antibiotics leads to the emergence of more drug-resistant strains, reducing the effectiveness of infected wound treatment.5–7 Consequently, new strategies with less drug resistance for antibacterial therapy are indispensable.Taking advantage of the potent broad-spectrum antibacterial activity, unique antibacterial mechanisms, and relatively weak induced drug resistance, antimicrobial peptides (AMPs) have become a good candidate to replace antibiotics.8–11 Nevertheless, some AMPs are limited by their poor stability under physiological conditions, and strong toxicity to eukaryotic cells (e.g., inducing hemolysis).12,13 To overcome these problems, numerous strategies have been developed to improve the stability of AMPs and to attenuate their toxicity to host cells, such as the optimization of their chemical structures and the loading of these peptides by using carriers.14–16
The rapid development of nanotechnology also provides promising methods for treating antibiotic-resistant bacteria. Through rational design, nanomaterials with an adjustable size, surface modification of drugs or functional groups, and stimulus responsiveness (drug release, generation of heat, reactive oxygen species, etc.) can be obtained. These materials either serve as carriers for the targeted delivery of antibiotics or have their own antibacterial properties.17–20 For example, through vacancy engineering and facet engineering, our group and co-workers synthesized a series of metal sulfide-based nanomaterials with photothermal and photodynamic antibacterial activities.21–23
Metal–organic frameworks (MOFs) are a class of porous nanomaterials composed of metal ions and organic ligands and also hold great promise for antibacterial therapy.24–27 On the one hand, MOFs exhibit high loading capacity for drugs (including antibiotics, photosensitizers, and photothermal molecules) due to their micro-porosity and easy functionalization. Additionally, some drugs also act as organic linkers. On the other hand, the released metal ions in MOFs could increase the antibacterial activity of MOFs.
Gallium (Ga), a kind of group IIIA metal, has an iron-like ionic radius but is redox-inactive.28 As an iron mimetic, Ga displays antimicrobial and anti-tumor activities as it can interfere in iron homeostasis in tumor, bacterial, or fungal cells, leading to a reduction of ribonucleotide reductase, mitochondrial dysfunction, and changes in proteins of iron transport and storage.29–31 In particular, as an FDA approved drug, the antibacterial mechanism, effect, and application of gallium nitrate have been extensively studied.32–35 Although there have been many studies on antibacterial MOFs, few studies on gallium-based MOFs have been reported.36 In this study, we synthesized a Ga-based MOF as a platform to load melittin (MEL), a typical AMP stemmed from honeybee (Apis mellifera) venom, and investigated its antibacterial activity against clinical isolated methicillin-resistant Staphylococcus aureus (MRSA) and Escherichia coli (E. coli). The MEL-loaded MOF (MM) was synthesized under mild pH and room temperature conditions. Importantly, the Ga ion and MEL can achieve synergistic antibacterial activity, reducing the side effects of MEL and can be used to treat wound infections.
2. Experimental
2.1. Materials
CTAB was bought from Biotopped. Gallium nitrate hydrate was bought from Shanghai Dibai Biotechnology. Disodium terephthalate (Na2BDC) was bought from Energy Chemical. MEL and fluorescein isothiocyanate (FITC)-MEL were synthesized by ChinaPeptides, China. MRSA (NKC281) was isolated from the Tianjin Third Central Hospital, Tianjin, China. E. coli is DH5α cells bought from Beijing Dingguo Changsheng Biotechnology.2.2. Synthesis of MOFC and MM
Ga(NO3)3 (100 mg) and Na2BDC (84 mg) were dissolved in 10 mL of H2O, respectively. Then, CTAB (40 mg) was dissolved in the Na2BDC solution and stirred for 5 min. Finally, the Ga(NO3)3 solution was added using an injection pump at 5 mL min−1, followed by magnetic stirring for 1 h. A white precipitate (MOFC) was obtained after the mixture was centrifuged (5000 rpm, 3 min). The white precipitate was then washed with H2O 3 times and collected by centrifugation. The obtained MOFC was dispersed in H2O and stored at 4 °C for further experiments.For the synthesis of MM, 10 mL of MEL was added into the Na2BDC–CTAB mixture with a final concentration of 1 mg mL−1. Then, Ga(NO3)3 solution (1 mg mL−1) with the same volume was added using an injection pump at 5 mL min−1, followed by magnetic stirring for 1 h. A white precipitate was collected by centrifugation and washed three times with H2O.
The characterization of the nanocomposites was mainly performed using a scanning electron microscope (SEM, TESCAN MIRA LMS, Czech), an X-ray diffractometer (XRD, SmartLab-SE, Japan), and a nanoparticle size/potentiometer (DLS and zeta potential, Malvern Panalytical, Zetasizer Nano ZS, UK).
The degradation rate of MOFC was determined by the concentration of Ga ions in the supernatant, which was measured using an inductively coupled plasma optical emission spectrometer (ICP-OES, Agilent 5110, USA). The CTAB residual in the final product was determined by testing the Br concentration using an inductively coupled plasma mass spectrometer (ICP-MS, Agilent 7800, USA).
2.3. MEL-loading/releasing assays
To measure the MEL-loading capacity, FITC labeled MEL (FITC-MEL) was used to prepare MM. Then, the contents of FITC-MEL in the supernatants and washing solutions were analysed using a fluorescence microplate reader (PerkinElmer, USA) to quantify the concentration of FITC-MEL. The MEL-loading capacity was calculated by (the FITC-MEL content of the initial solution – the content in the supernatant – the content in the washing solution)/the content of nanoparticles × 100%.15The MEL-releasing capacity of MM was detected in PBS (pH 7.4) and MES (pH 6.0) buffers, respectively. After incubation for 8, 16, 24, and 48 h, the suspensions were sampled and centrifuged at 8000 rpm for 5 min. The FITC-MEL concentration of the supernatants was quantified using a fluorescence microplate reader. The MEL releasing capacity of MM was calculated using the FITC-MEL content in the supernatant/the initial FITC-MEL content × 100%.
2.4. Antibacterial activity of MM
The MRSA strain NKC281 was isolated from the Third Central Hospital of Tianjin and stored in the Laboratory of Modern Mycology. The strain was cultured overnight at 37 °C and resuspended to OD600 = 0.01. Then, 100 μL of the cell suspension with different concentrations of MOFC, MEL, Ga ions and MM was added in 96-well plates and cultured at 37 °C for 12 h. The final OD600 was then tested. The suspension was also diluted 1000 times and the number of viable MRSA was tested by colony-forming unit (CFU) assays on LB solid plates.For E. coli, the strain was cultured overnight at 37 °C and resuspended to OD600 = 0.01. Then, 100 μL of the cell suspension with different concentrations of MEL, Ga ions and MM was added in 96-well plates and cultured at 37 °C for 12 h. The final OD600 was then tested.
For SEM observation, 100 μL of MRSA in LB liquid medium (OD600 = 1) was mixed with PBS, MOFC, MEL, MM, Ga ion (10 ppm), Ga ion (150 ppm), and Ga ion + MEL (10 + 10 ppm), respectively. The mixture was then plated on a piece of cover glass, and the cells were fixed by 3% formaldehyde for 2 h, dehydrated by ethanol solutions (30, 50, 70, 90 and 100% of volume fraction, respectively), dried using a freeze drier (10N, SCIENTZ, China), and observed using a SEM.
2.5. Membrane potential tests
Membrane potential changes in the bacteria with different treatments were detected using a BacLight Bacterial Membrane Potential Kit (Invitrogen). Initially, The MRSA cells (OD600 = 0.5) were incubated with PBS (set as the control group), MOFC, MEL, and MM, respectively for 2 h. The bacteria were then collected, washed, and re-dispersed in PBS. Bis-(1,3-dibutylbarbituric acid) trimethine oxonol (DiBAC4(3)) was added to all samples at a final concentration of 8 μM and stained for 15 min. The fluorescence intensity of the samples was evaluated using a flow cytometer (Guava easyCyte6-2L, USA).2.6. ROS production tests
100 μL of MRSA suspensions (OD600 = 0.5) was incubated with MOFC, MEL, MM and PBS at 37 °C in a 96-well microplate for 2 h. Then, 1 μM 2′,7′-dichlorofluorescin diacetate (DCFH-DA) was added. The cells were further incubated for 0.5 h. The fluorescence intensity of the cells was measured using a fluorescence microplate reader (excitation wavelength = 488 nm and emission wavelength = 520 nm).2.7. Cytotoxicity assay
The DC2.4 cells were cultured in DMEM medium including 10% (v/v) fetal bovine serum (Gibco), penicillin (100 U mL−1), and streptomycin (100 U mL−1) in a humidified atmosphere of 5% CO2 at 37 °C. 1 × 104 cells were then seeded in a 96-well plate and incubated 24 h. Then different amounts of MM, MOFC, and MEL were added into the cell cultures, and the cells were incubated for another 24 h. The cell viability was detected using the CCK-8 assay kit (Solarbio, China). The apoptosis of cells was also evaluated using a flow cytometer (Guava easyCyte6-2L, USA).2.8. In vivo wound healing
The wound healing efficiency in vivo was evaluated using a mouse skin-infection model. The animal experiments were approved by the Animal Care and Use Committee at Nankai University (Approval number 2023-SYDWLL-000367). The wound of mice was infected with 100 μL of MRSA (1 × 108 cells per mL), and smeared with 100 μL of PBS, MOFC, MEL and MM (0.1 mg mL−1) per day for three times, respectively. The wounds were observed for 7 days for photographing and sampling. The wounds with surrounding tissues were collected and homogenized in PBS, and the number of MRSA was tested using CFU assays in solid LB plates.2.9. Histological evaluation
The collected wound tissues as well as the main organs of mice (i.e., heart, liver, spleen, lungs, and kidneys) were fixed with 4% formaldehyde, embedded in paraffin, and stained by hematoxylin and eosin (H&E) for histopathological observation.2.10. Cytokine expression
The homogenized wound tissues were centrifuged, and the levels of IL-6 and TNF-α in supernatants were tested using the corresponding ELISA kits (Jianglaibio) based on the manufacturer's instructions.2.11. Statistical analysis
Each experiment was performed in triplicate. The data were described as mean ± standard deviation (SD). All statistical analyses were performed using the ANOVA test (P < 0.05) using the SPSS software (Version 22, IBM, USA).3. Results and discussion
3.1. Synthesis and characterization of the nanocomposites
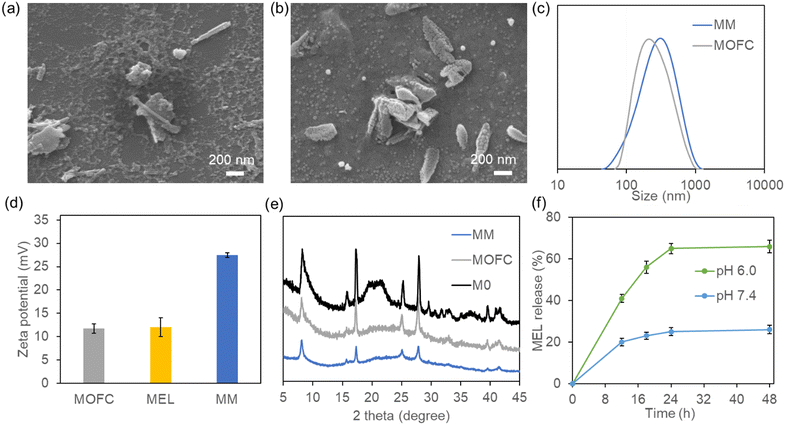
Traditionally, Ga-based MOFs were prepared via the solvothermal method using autoclaves at high temperature, making it difficult to achieve one-step drug loading.37 To solve this problem, we changed the organic ligand from 1,4-dicarboxybenzene to disodium terephthalate (Na2BDC),38 and Ga-based MOFs could be easily synthesized by simply mixing Na2BDC and Ga(NO3)3 at room temperature in the aqueous phase (M0). In order to reduce the size of the MOF and improve its stability, we further introduced the surfactant CTAB during synthesis.39,40The scanning electron microscopy (SEM) results showed that the synthesized M0 appeared as rod-like crystallites of 1–2 μm length (Fig. S1, ESI†), which was similar to our previously synthesized MIL-68(Ga).37 In the presence of CTAB, the obtained MOFC became an irregular shape, with some short and sharper rod like particles, and the size was less than 1 μm (Fig. 1a). With the addition of MEL, MM exhibited jujube stone-like shape, and the size and shape of MM were more uniform compared with those of MOFC (Fig. 1b). The change of size was further confirmed by DLS (Fig. S2a and Fig. 1c, ESI†). Considering that MEL can also act as the positive surfactant, the addition of MEL could also regulate the shape of MM and reduce its size. The zeta potential results showed that M0 is nearly neutral (2.5 mV, Fig. S2b, ESI†), while MOFC and MEL are positive (about 12 mV). The MEL loaded MOFC, i.e., MM, was much more positive (27 mV, Fig. 1d). The interaction between MEL and MOFC may change their conformation, leading to the exposure of more positive groups and consequently higher zeta potential of the obtained MM than single MEL or MOFC. Moreover, the XRD patterns of M0, MOFC and MM implied the existence of crystalline structure, which is consistent with our previous results (Fig. 1e).36,37
The MEL loading capacity of MOFC was obviously higher than that of our previously reported MOF-MEL nanocomposite as about 90% of MEL was loaded in the MM,15 and the final loading capacity was 45%. To investigate the MEL release capacity, the as-prepared MM was incubated in PBS buffer (pH 7.4) and MES buffer (pH 6.0), respectively. The release amount of FITC-MEL was tested at different time nodes and the results are shown in Fig. 1f. The release capacity of MEL was more potent at pH 6.0 than pH 7.4, which was attributed to the pH sensitivity of MM. Moreover, the stabilities of MOFC and MM in the aqueous solution were evaluated. After incubation at pH 6.0 and pH 7.4 for 12 h, the solution was centrifuged and the amount of Ga in the supernatant was tested using an inductively coupled plasma optical emission spectrometer (ICP-OES). As shown in Fig. S3 (ESI†), ∼30% of MM and 36% of MOFC were degraded at pH 6.0, while 6% of MM and 8% of MOFC were degraded at pH 7.4. To see whether there was CTAB residual in MM, we tested the bromine (Br) content of MM after cleaning up by ICP-MS, and the results showed that neglectable CTAB existed in MM (<1 μg per mg MM).
3.2. Antibacterial activity of MM
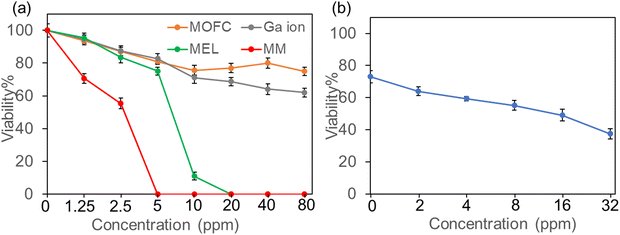
As both Ga and MEL have been demonstrated with antibacterial activity, we then evaluated the antibacterial activity of MM on the growth of drug resistance strains, MRSA (isolated from clinic). MRSA was incubated with four treatments (MOFC, Ga(NO3)3, MEL, and MM) at a series of concentrations (0–80 ppm) for 12 h, respectively, and the OD600 value was measured to evaluate the effect of concentration on antibacterial activity. As shown in Fig. 2a, compared with MOFC, Ga(NO3)3 exhibited moderate antibacterial activity in the tested concentration. This is acceptable as Ga(NO3)3 has antibacterial properties but requires a relatively high concentration.34 Noteworthily, less than 10% of bacterial cells survived with 10 ppm of MEL, indicating the strong antibacterial activity of MEL. More strikingly, nearly no bacterial cells survived in the presence of 5 ppm MM, demonstrating that MM exhibits the best antibacterial activity. As MOFC can be degraded to release Ga3+, we then tested the synergy effects of Ga3+ and MEL. As shown in Fig. 2b, the addition of Ga(NO3)3 could enhance the antibacterial effect of MEL obviously, suggesting that the release of MEL and Ga3+ in MM plays a key role in the potent antibacterial activity of MM.The antibacterial activity of the nanocomposites against MRSA was also verified by counting live cells on agar plates. MRSA was incubated with different drugs at the same concentration (10 ppm) for 2 hours, and then plated on LB solid plates for 24 h. As shown in Fig. S4 (ESI†), the comparison of bacterial colonies growing on the plates demonstrated that the antibacterial activity of MM was much stronger than other agents.
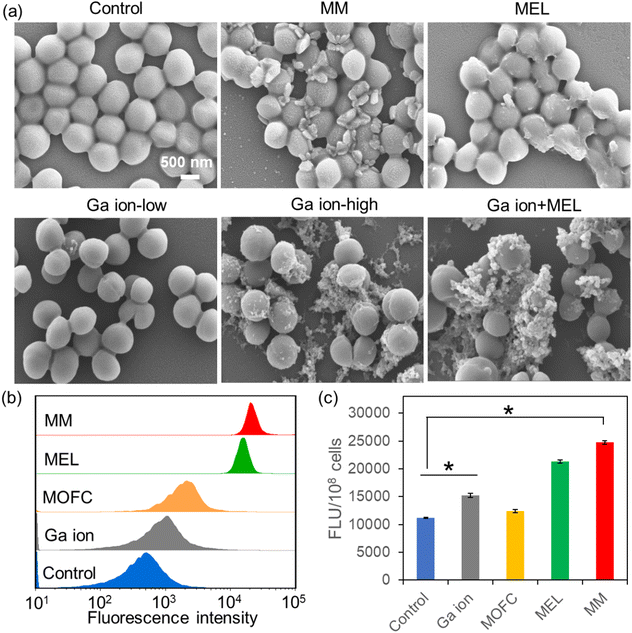
Then, we carried out SEM experiments to observe the morphological changes of MRSA after different treatments (Fig. 3a). Compared with the untreated MRSA, many large cell fragments could be observed in the MM treated group, while in the MEL-treated group, only several cells were destroyed completely. Moreover, the antibacterial activity of Ga ions is concentration-dependent, as the low concentration of Ga ions could not kill MRSA, while a high amount of Ga ions could effectively kill MRSA. Interestingly, MEL with a low concentration of Ga ions exhibited much stronger antibacterial activity than MEL at the same concentration (10 ppm). In order to confirm that the fragments were broken bacterial cells rather than the MOF materials, energy dispersive spectroscopy (EDS) analysis for C, N, O, Ga elements of the Ga ion + MEL group was conducted. As shown in Fig. S5 (ESI†), there was almost no Ga, considering that when the Ga ions and MEL in combination were added to the bacterial cultures, the fragments in SEM all belonged to MRSA, suggesting that high concentrations of Ga ions as well as Ga ions + MEL could also effectively kill MRSA.
Cell membrane depolarization is a well-established mechanism for antibacterial agents.41 The membrane potential of MRSA after different treatments (10 ppm) was then tested by staining of DiBAC4(3), a specific probe that can enter depolarized cells, leading to enhanced fluorescence.42 As shown in Fig. 3b, MOFC and Ga(NO3)3 could change the membrane potential of MRSA a little. However, MRSA treated with MEL and MM exhibited a prominent enhancement of membrane potential, suggesting that MEL and MM could induce bacterial inactivation via a membrane depolarization mechanism.43
Another antibacterial mechanism is inducing oxidative damage through reactive oxygen species (ROS) generation by drugs.44 The effects of Ga(NO3)3, MOFC, MEL and MM on bacterial ROS generation were then investigated. As shown in Fig. 3c, at 10 ppm of different agents, Ga(NO3)3 promoted ROS production compared to MOFC. This phenomena can be explained by recent studies, i.e., Ga(NO3)3 could induce the intracellular accumulation of ROS.33,45,46 Meanwhile, MM has the most significant effect on enhancing bacterial ROS levels, leading to the greatest damage to cells.
Apart from Gram-positive bacteria, we also tested the antibacterial activity of MM on Escherichia coli (E. coli), a typical Gram-negative bacterium. As shown in Fig. S6 (ESI†), both MEL and MM exhibit antibacterial activity toward E. coli, but the concentration needed was higher than MRSA, which was consistent with the previously reported results, indicating a greater sensitivity of Gram-positive bacteria to MEL compared with Gram-negative bacteria.47
3.3. MM does not exhibit cytotoxicity to DC2.4 cells
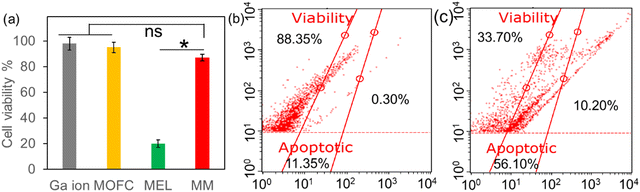
Though exhibiting potent antibacterial activity, the cytotoxicity towards mammalian cells at therapeutic doses should also be considered. The cytotoxicity of the agents was then assessed via the CCK-8 method. After incubation with 20 ppm of different agents at 37 °C for 24 h, the viability of DC2.4 cells was examined. As shown in Fig. 4a, the relative cell viability was more than 80% after Ga(NO3)3, MOFC and MM treatments, in favour of the potential in vivo application for MM. However, only about 20% of cells survived in the presence of MEL, thus limiting the clinical application of MEL. The cytotoxicity of MM and MEL was also compared through flow cytometry. As shown in Fig. 4b and c, after 12 h incubation of MEL, 10.2% of DC2.4 cells were dead and 56.1% of cells were apoptotic. In contrast, after incubation of MM, the cell viability was 88.35%, further indicating that MM exhibited much less cytotoxicity towards DC2.4 cells compared to MEL. Based on the MEL loading capacity of MM, the MEL concentration in the MM group was less than 10 ppm, which was a key reason for less cytotoxicity compared with the MEL group.3.4. In vivo antibacterial activity of MM
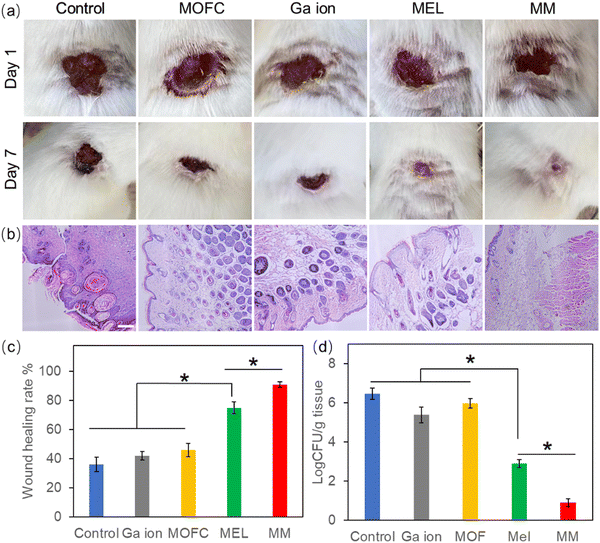
Based on the excellent antibacterial results of MM in vitro, the MRSA-infected mouse model was used to test the wound healing ability in vivo.48 The wounds on the backs of mice were infected by MRSA and then treated with PBS (control), MOFC, Ga(NO3)3, MEL and MM, respectively. After 7 days, the wound healing rate of the control group was less than 40%. MEL improved wound healing to some extent, while the MM treated group exhibited the highest healing rate (above 90%), demonstrating that MM effectively promoted wound healing against bacterial infections (Fig. 5a and c). Consistently, histological analysis further showed that the MM treated group exhibited the best tissue regeneration ability (Fig. 5b). Moreover, the MM treated group exhibited the lowest level of bacteria on the wound tissue than other groups (Fig. 5d). Additionally, the histopathological analysis of the main tissues, including heart, liver, spleen, lungs and kidneys, for the MM group and that for the control group were compared. As shown in Fig. S7 (ESI†), there was no histological damage or abnormality between two groups, demonstrating the good biosafety of MM.Inflammatory response is a defensive phenomenon in the human body. The expression levels of inflammatory cytokines, including IL-6 and TNF-α, were tested to evaluate the healing effect of MM.49,50 Excessive amount of IL-6 can bring pain and affect the immune system.51 As shown in Fig. 6a, all agent treated groups exhibited a reduced amount of IL-6, and the IL-6 level of the Ga(NO3)3-treated group was lower than the groups treated with MEL or MOFC, which was attributed to the anti-inflammatory effects of Ga.52 Strikingly, the IL-6 levels of the MM-treated group was much lower than those of other groups, demonstrating that MM strongly reduces IL-6 production to inhibit inflammation for rapid wound healing.
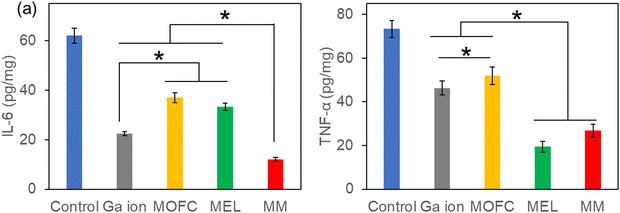 | ||
| Fig. 6 IL-6 (a) and TNF-α (b) expression in the wounds with different treatments. The asterisks indicate a significant difference between the groups (P < 0.05). | ||
Excessive expression of TNF-α is also detrimental to wound healing as it may inhibit cell migration and collagen deposition and even damage microvessels.53 As shown in Fig. 6b, the TNF-α levels of the mice treated with MM or MEL were also much lower than those of the control group and the groups treated with Ga(NO3)3 or MOFC. Together, these results revealed that MM significantly downregulated the production of the inflammatory cytokines IL-6 and TNF-α, which accelerated wound healing.
4. Conclusions
In conclusion, this study developed a novel gallium-based MOF platform for loading AMPs for the synergistic inhibition of drug-resistant bacteria and wound healing. Gallium-based MOFs, which were synthesized at mild room temperature, efficiently loaded the model antimicrobial peptide MEL, forming nanocomposites MM with high antibacterial activity and good biocompatibility to mammalian cells. Notably, the obtained nanocomposites showed much higher antibacterial activity against methicillin-resistant Staphylococcus aureus and Escherichia coli as compared to MEL and gallium ions. The MM nanocomposites further accelerated wound healing via down regulation of the inflammatory cytokines IL-6 and TNF-α. Owing to the potent antibacterial activity, good biocompatibility and easy preparation, the MM nanocomposites can serve as an excellent candidate for treating antibiotic-resistant bacteria.Author contributions
Shuo Liu: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft, and writing – review and editing; Yuxin Ji: methodology and drawing; Hangqi Zhu: writing – review and editing; Zhishang Shi: methodology; Mingchun Li: conceptualization and writing – review and editing; Qilin Yu: conceptualization, funding acquisition, project administration, resources, and supervision. All the authors revised the manuscript and approved it in the present form.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China, China (32170102), the Natural Science Foundation of Tianjin (21JCYBJC01420), and the Fundamental Research Funds for the Central Universities (63233050). The authors would also like to thank the Shiyanjia Lab (Available online: www. shiyanjia.com accessed on 11 June 2023) for the SEM, XRD and ICP-MS analyses.References
- A. Y. Peleg and D. C. Hooper, New Eng. J. Med., 2010, 362, 1804–1813 CrossRef CAS PubMed.
- Z. Wang, F. Qi, H. Luo, G. Xu and D. Wang, Front. Immunol., 2022, 13, 789274 CrossRef CAS PubMed.
- C. K. Sen, Adv. Wound Care, 2021, 10, 281–292 CrossRef PubMed.
- A. K. Dabrowska, F. Spano, S. Derler, C. Adlhart, N. D. Spencer and R. M. Rossi, Skin Res. Technol., 2018, 24, 165–174 CrossRef CAS PubMed.
- E. M. Darby, E. Trampari, P. Siasat, M. S. Gaya, I. Alav, M. A. Webber and J. M. A. Blair, Nat. Rev. Microbiol., 2023, 21, 280–295 CrossRef CAS PubMed.
- R. Urban-Chmiel, A. Marek, D. Stepien-Pysniak, K. Wieczorek, M. Dec, A. Nowaczek and J. Osek, Antibiotics, 2022, 11, 1079 CrossRef CAS PubMed.
- M. Denk-Lobnig and K. B. Wood, Curr. Opin. Microbiol., 2023, 74, 102306 CrossRef CAS PubMed.
- J. Xuan, W. Feng, J. Wang, R. Wang, B. Zhang, L. Bo, Z. S. Chen, H. Yang and L. Sun, Drug Res. Updates, 2023, 68, 100954 CrossRef CAS PubMed.
- X. Chen, J. Han, X. Cai and S. Wang, Biotechnol. Adv., 2022, 60, 108012 CrossRef CAS PubMed.
- G. Li, Z. Lai and A. Shan, Adv. Sci., 2023, 10, 2206602 CrossRef CAS PubMed.
- Z. Lyu, P. Yang, J. Lei and J. Zhao, Antibiotics, 2023, 12, 1037 CrossRef CAS PubMed.
- K. B. S. Oliveira, M. L. Leite, V. A. Cunha, N. B. Cunha and O. L. Franco, Drug Discovery Today, 2023, 28, 103629 CrossRef PubMed.
- A. Mazurkiewicz-Pisarek, J. Baran and T. Ciach, Int. J. Mol. Sci., 2023, 24, 9031 CrossRef CAS PubMed.
- Z. Lai, X. Yuan, H. Chen, Y. Zhu, N. Dong and A. Shan, Biotechnol. Adv., 2022, 59, 107962 CrossRef CAS PubMed.
- D. Yu, Y. Wang, J. Zhang, Q. Yu, S. Liu and M. Li, J. Mater. Sci., 2022, 57, 16809–16819 CrossRef CAS.
- J. K. White, S. Mohanty, T. Muhammad, M. A. S. Campa, W. E. Houssen, N. Ferraz, U. Göransson and A. Brauner, ACS Infect. Dis., 2023, 9, 1056–1063 CrossRef CAS PubMed.
- M. Xie, M. Gao, Y. Yun, M. Malmsten, V. M. Rotello, R. Zboril, O. Akhavan, A. Kraskouski, J. Amalraj, X. Cai, J. Lu, H. Zheng and R. Li, Angew. Chem., Int. Ed., 2023, 62, e202217345 CrossRef CAS PubMed.
- J. He, M. Hong, W. Xie, Z. Chen, D. Chen and S. Xie, J. Controlled Release, 2022, 351, 301–323 CrossRef CAS PubMed.
- Z. Zhou, M. Kai, S. Wang, D. Wang, Y. Peng, Y. Yu, W. Gao and L. Zhang, WIREs Nanomed. Nanobiotechnol., 2023, 15, e1881 CrossRef CAS PubMed.
- S. Roy, I. Hasan and B. Guo, Coordin. Chem. Rev., 2023, 482, 215075 CrossRef CAS.
- J. Sun, J. Wen, J. Wang, Y. Yang, G. Wang, J. Liu, Q. Yu and M. Liu, J. Hazard. Mater., 2023, 451, 131082 CrossRef CAS PubMed.
- S. Mo, Y. Zhao, J. Wen, J. Sun, Z. Zhang, Q. Yu, G. Wang, X. Chen and M. Liu, J. Hazard. Mater., 2022, 432, 128662 CrossRef CAS PubMed.
- J. G. Kim, J. Sun, Y. Zhao, J. Wen, B. Zhou, Z. Zhang, S. Mo, J. Wang, H. Liu, G. Wang, Q. Yu and M. Liu, Small, 2022, 18, 2107807 CrossRef CAS PubMed.
- M. Moharramnejad, R. E. Malekshah, A. Ehsani, S. Gharanli, M. Shahi, S. A. Alvan, Z. Salariyeh, M. N. Azadani, J. Haribabu, Z. S. Basmenj, A. Khaleghian, H. Saremi, Z. Hassani and E. Momeni, Adv. Colloid Interfaces, 2023, 316, 102908 CrossRef CAS PubMed.
- D. Han, X. Liu and S. Wu, Chem. Soc. Rev., 2022, 51, 7138–7169 RSC.
- X. Qi, N. Shen, A. A. Othman, A. Mezentsev, A. Permyakova, Z. Yu, M. Lepoitevin, C. Serre and M. Durymanov, Pharmaceutics, 2023, 15, 1521 CrossRef CAS PubMed.
- S. Chen, J. Lu, T. You and D. Sun, Coordin. Chem. Rev., 2021, 439, 213929 CrossRef CAS.
- N. Kircheva and T. Dudev, Inorg. Chem., 2020, 59, 6242–6254 CrossRef CAS PubMed.
- N. Kircheva and T. Dudev, J. Inorg. Biochem., 2021, 214, 111309 CrossRef CAS PubMed.
- A. B. Kelson, M. Carnevali and V. Truong-Le, Curr. Opin. Pharmacol., 2013, 13, 707–716 CrossRef CAS PubMed.
- F. Kurtuldu, N. Mutlu, A. R. Boccaccini and D. Galusek, Bioact. Mater., 2022, 17, 125–146 CAS.
- L. Rossato, J. P. Arantes, S. M. Ribeiro and S. Simionatto, Diagn. Microbiol. Infect. Dis., 2022, 102, 115569 CrossRef CAS PubMed.
- M. Guo, P. Tian, Q. Li, B. Meng, Y. Ding, Y. Liu, Y. Li, L. Yu and J. Li, Microbiol. Spectrum, 2023, 11, e0033423 CrossRef PubMed.
- Z. Xu, X. Zhao, X. Chen, Z. Chen and Z. Xia, RSC Adv., 2017, 7, 52266–52273 RSC.
- F. Shamkani, S. M. Barzi, F. Badmasti, M. Chiani, E. Mirabzadeh, M. Zafari and M. Shafiei, Int. Immunopharmacol., 2023, 115, 109551 CrossRef CAS PubMed.
- J. Yang, C. Wang, X. Liu, Y. Yin, Y. H. Ma, Y. Gao, Y. Wang, Z. Lu and Y. Song, Adv. Funct. Mater., 2020, 30, 2004861 CrossRef CAS.
- Y. Zhang, L. Liu, D. Yu, J. Liu, L. Zhao, J. Liu and S. Liu, Molecules, 2022, 27, 3443 CrossRef CAS PubMed.
- X. Cheng, S. Zhang, H. Liu, H. Chen, J. Zhou, Z. Chen, X. Zhou, Z. Xie, Q. Kuang and L. Zheng, ACS Appl. Mater. Interfaces, 2020, 12, 36996–37005 CrossRef CAS PubMed.
- J. Zhao, X. Liu, Y. Wu, D. S. Li and Q. Zhang, Coordin. Chem. Rev., 2019, 391, 30–43 CrossRef CAS.
- L. Zhang, J. Xian, X. Xiang, H. Ouyang, L. Wang and Z. Fu, Sens. Actuators, B, 2022, 373, 132774 CrossRef CAS.
- T. Huang, J. A. Holden, E. C. Reynolds, D. E. Heath, N. M. O’Brien-Simpson and A. J. O’Connor, ACS Appl. Mater. Interfaces, 2020, 12, 55696–55709 CrossRef CAS PubMed.
- M. S. Kwun and D. G. Lee, Life Sci., 2021, 285, 120003 CrossRef CAS PubMed.
- S. F. A. Albaayit, R. Maharjan, R. Abdullah and M. H. M. Noor, J. Appl. Biomed., 2022, 20, 15–21 CrossRef PubMed.
- X. Wang, H. Wang, J. Cheng, H. Li, X. Wu, D. Zhang, X. Shi, J. Zhang, N. Han and Y. Chen, Chem. Eng. J., 2023, 466, 143201 CrossRef CAS.
- L. Li, H. Chang, N. Yong, M. Li, Y. Hou and W. Rao, J. Mater. Chem. B, 2021, 9, 85–93 RSC.
- Z. W. Scott, S. R. Choi, G. A. Talmon, B. E. Britigan and P. Narayanasamy, ACS Infect. Dis., 2022, 8, 2096–2105 CrossRef CAS PubMed.
- T. Picoli, C. M. Peter, J. L. Zani, S. B. Waller, M. G. Lopes, K. N. Boesche, G. D. Vargas, S. O. Hübner and G. Fischer, Microb. Pathog., 2017, 112, 57–62 CrossRef CAS PubMed.
- L. Peng, H. Wei, L. Tian, J. Xu, M. Li and Q. Yu, Sci. China Mater., 2021, 64, 759–768 CrossRef CAS.
- S. Lin, L. Pei, W. Zhang, G. Shu, J. Lin, H. Li, F. Xu, H. Tang, G. Peng, L. Zhao, L. Yin, L. Zhang, R. Huang, S. Chen, Z. Yuan and H. Fu, Mater. Sci. Eng., C, 2021, 130, 112450 CrossRef CAS PubMed.
- C. Li, C. Chen, J. Zhao, M. Tan, S. Zhai, Y. Wei, L. Wang and T. Dai, ACS Biomater. Sci. Eng., 2021, 7, 3898–3907 CrossRef CAS PubMed.
- S. Ronchetti, G. Migliorati and D. V. Delfino, Biomed. Pharmacother., 2017, 96, 1445–1452 CrossRef CAS PubMed.
- V. A. W. Sales, T. R. R. Timóteo, N. M. Silva, C. G. Melo, A. S. Ferreira, M. V. G. Oliveira, E. O. Silva, L. M. S. Mendes, L. A. Rolim and P. J. R. Neto, Curr. Med. Chem., 2021, 28, 2062–2076 CrossRef PubMed.
- Q. C. Chen, J. Wu, Y. Liu, Y. Q. Li, C. Q. Zhang, W. C. Qi, K. W. K. Yeung, T. M. Wong, X. L. Zhao and H. B. Pan, Mater. Sci. Eng., C, 2019, 105, 110083 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3tb01754k |
| This journal is © The Royal Society of Chemistry 2023 |