Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Surface-enhanced Raman spectroscopy for bioanalysis and diagnosis
Muhammad Ali
Tahir†
a,
Nicoleta E.
Dina†
 *b,
Hanyun
Cheng
a,
Ventsislav K.
Valev
*b,
Hanyun
Cheng
a,
Ventsislav K.
Valev
 *c and
Liwu
Zhang
*c and
Liwu
Zhang
 *ad
*ad
aShanghai Key Laboratory of Atmospheric Particle Pollution and Prevention, Department of Environmental Science & Engineering, Fudan University, Shanghai, 200433, Peoples’ Republic of China. E-mail: zhanglw@fudan.edu.cn
bDepartment of Molecular and Biomolecular Physics, National Institute for Research and Development of Isotopic and Molecular Technologies, 67-103 Donat, 400293 Cluj-Napoca, Romania. E-mail: nicoleta.dina@itim-cj.ro
cCentre for Photonics and Photonic Materials, Department of Physics, University of Bath, Bath, BA2 7AY, UK. E-mail: vkv23@bath.ac.uk
dShanghai Institute of Pollution Control and Ecological Security, Shanghai, 200092, Peoples’ Republic of China
First published on 6th July 2021
Abstract
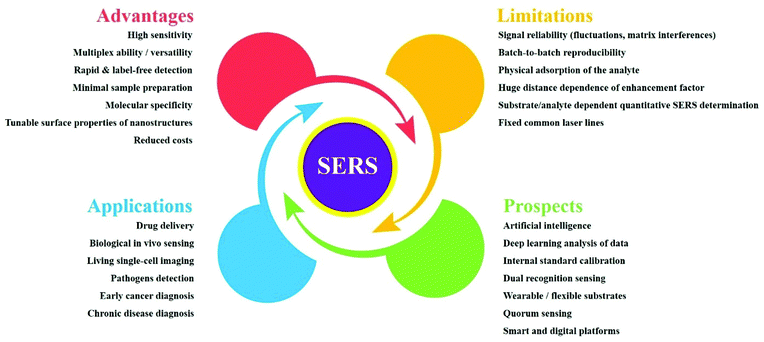
In recent years, bioanalytical surface-enhanced Raman spectroscopy (SERS) has blossomed into a fast-growing research area. Owing to its high sensitivity and outstanding multiplexing ability, SERS is an effective analytical technique that has excellent potential in bioanalysis and diagnosis, as demonstrated by its increasing applications in vivo. SERS allows the rapid detection of molecular species based on direct and indirect strategies. Because it benefits from the tunable surface properties of nanostructures, it finds a broad range of applications with clinical relevance, such as biological sensing, drug delivery and live cell imaging assays. Of particular interest are early-stage-cancer detection and the fast detection of pathogens. Here, we present a comprehensive survey of SERS-based assays, from basic considerations to bioanalytical applications. Our main focus is on SERS-based pathogen detection methods as point-of-care solutions for early bacterial infection detection and chronic disease diagnosis. Additionally, various promising in vivo applications of SERS are surveyed. Furthermore, we provide a brief outlook of recent endeavours and we discuss future prospects and limitations for SERS, as a reliable approach for rapid and sensitive bioanalysis and diagnosis.
1. Introduction
In applied sciences, rapid, accurate and sensitive response are key aspects of all analytical techniques; this is especially true when it comes to detection and therapeutic issues within the healthcare environment. For reliable disease diagnosis and quick therapeutic actions, it is mandatory to have a clear understanding of biological pathways and processes. Moreover, it is essential to have specific and ultrasensitive analytical tools for identifying clinically relevant molecular species. Traditional methods for bioanalysis and disease diagnosis are time consuming, as they require series of biochemical tests. Such delay is incompatible with medical emergencies. Currently, there are different analytical tools, such as fluorescence immunoassay, enzyme-linked immunosorbent assay (ELISA), radio immunoassay, fluorescence spectroscopy, mass spectrometry, etc. that are developed for biomolecule detection and biomedical imaging, based on biological, physical and chemical phenomena. However, these tools are time consuming, they need favourable conditions for analysis and expensive protocols, and sometimes they can struggle to solve real-life practical issues (e.g. being unable to discriminate between live and dead pathogens). For public health safety and disease prevention, there is a dire need for detection methods that offer high sensitivity, reproducibility, fast sample preparation time, user-friendliness and cost-effectiveness. This need covers a wide range of biomedical applications including vibrational spectroscopy for pathologies with reliable, fit-to-purposes methods for bioanalysis and diagnosis.Benefiting from the recent advancements in optics, laser technology and nano-devices, surface-enhanced Raman scattering (SERS) has become a widely used spectroscopic technique. The reasons are very straightforward: it can provide structural information at the single-molecule level1–4 owing to the extremely large electromagnetic field amplification that is produced by the excitation of localized surface plasmons (LSPs). Moreover, SERS is non-invasive, oblivious to water (omnipresent in biological samples) and can probe conformational changes, as it provides vibrational information on the targeted bioanalytes.5 Several reviews show the potential of SERS in therapeutic drug monitoring (TDM),6 in detecting cancer cells and tumor margins in vivo conditions7 or in DNA biosensing.8 SERS could replace standard methods that are currently routinely used for DNA-based diagnosis. The possibility to enhance disease diagnosis by employing SERS-based applications to detect biological markers at trace levels was evaluated recently in a comprehensive review by Chisanga et al.9 The current requirements of point-of-care (PoC) biosensing, during an outbreak of a very contagious virus, are related more than ever to strict biosafety regulations when transporting patients or biological samples to laboratory premises. The safe choice of portable handheld Raman devices with miniaturized, reproducible enhancing substrates as sample support is becoming increasingly necessary. In practice, rapid sampling, on a large scale, as a daily routine is met by the amplified Raman signals, provided by SERS, which facilitate fast trace analysis of clinically relevant biomolecules in ambient conditions, involving minor sample preparation. For instance, SERS-based detection of bacteria is fast and can be performed at the single-cell level. It could be used for diagnosing human infections (e.g. with antibiotic-resistant bacteria) using culture-free methods, directly from patient fluids or even in vivo.10 SERS also benefits from recent efforts to develop tandem microfluidic devices in order to improve in situ bioanalysis with direct clinical applicability.7
SERS is regarded as an ultrasensitive technique because it can achieve single-molecule (SM) detection. In large part, this sensitivity is due to the enhancement (by factors up to 108 or even larger) of inelastic light scattering by those molecules that are adsorbed on the surface of nanostructured metals, such as gold or silver nanoparticles (NPs).11–13 SERS also offers structural information in biological media in ambient environments.14 SERS surface selection rules mandate that the intensity from vibrational modes perpendicular to the surface is increased while maintaining that from the parallel modes constant.12,15 Several environmental parameters, such as pH or temperature, can affect the surface adsorption of the bioanalytes, and thus can influence also their spectral fingerprint.16
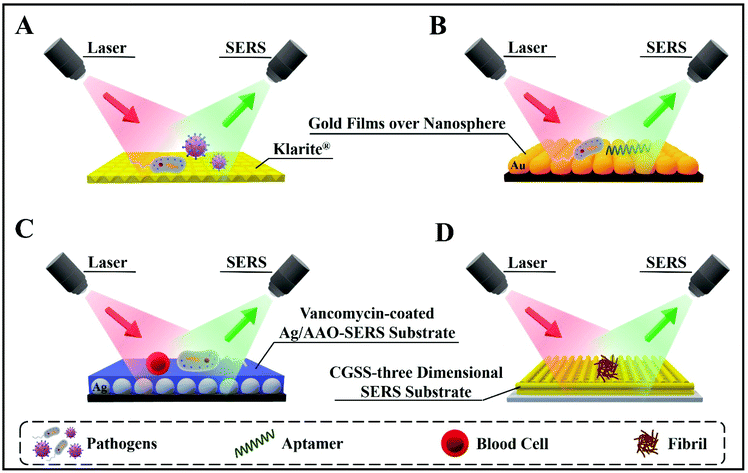
Importantly, SERS is user-friendly, compared with other highly sensitive techniques (such as mass spectrometry), and is applicable to aqueous solutions without the need for complex sample preparation protocols. Overall, SERS is quickly becoming an analytical technique of choice for successful applications in bioanalysis, disease detection and diagnosis.9,17 Furthermore, SERS enables disease diagnosis in vivo, i.e. inside living organisms, after coating biocompatible SERS-active NPs or substrates with appropriate surfactants and/or protective shells.18Fig. 1 presents representative examples of SERS-based bioassays reported in previous literature for detecting bioanalytes directly or via specific receptors, aptamers, etc. The versatility of SERS-based detection techniques is highlighted by showing different SERS substrates designed for detecting whole cells or clinically relevant bioanalytes with precise accuracy and high sensitivity.
There are two main approaches when using SERS for bioanalyte detection, namely label-based detection with SERS tags and label-free SERS detection (Fig. 2).19 SERS label-based detection aims to recognize the vibrational spectroscopic fingerprints of bioanalytes via indirect interaction, by using Raman reporter molecules to label the SERS tags; for instance, this detection is widely used in SERS-based DNA sensing.20–22 Label-free SERS detection is a direct approach; it senses and images the bioanalytes after adsorption on the SERS substrates/nanostructures, which often results in improved signal intensity.23 In the label-based detection scheme, the SERS nano-substrates comprise four distinct parts: (1) a nanostructured metallic substrate, usually made of gold or silver to improve the SERS intensity and activity; (2) an appropriate protective shell or layering to enhance the stability and biocompatibility; (3) a Raman reporter molecule layer to enable label-based detection with a unique SERS fingerprint; and (4) target-specific detection using bioconjugations. In the label-free detection scheme, the third part is omitted.
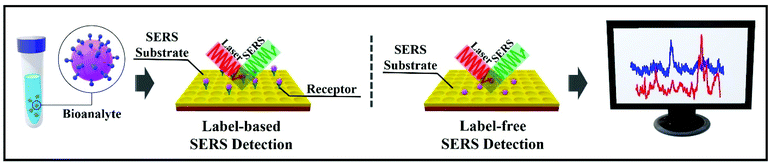 | ||
| Fig. 2 Diagrams of the two SERS-based approaches for bioanalyte detection in spectral pathology. On the left, label-based SERS detection. On the right, label-free SERS detection. | ||
A number of challenges remain for further applicability of SERS in biological and clinical environments. For instance, signal reliability can be an issue. Importantly, SERS has the potential to tackle common issues that are challenging for other bioanalytical techniques. Whether the technique lives up to its potential will depend on elucidating a number of questions, such as:
1. What nature of bioanalytes can be detected by SERS? What exactly are the practical limitations of direct and indirect SERS detection?
2. How do bioanalytes act after interacting with the nanoparticles? Do the SERS analyses affect the biological processes during in vivo detection?
3. Do the biological conditions alter the sensing properties of nanoparticles?
4. How effective is SERS for rapid and sensitive in vivo diagnosis?
For the successful application of SERS in both the label-based and label-free approaches, it is important to optimize the optical response of the plasmonic nanostructures. As there is a plethora of plasmonic nanostructures involved in various SERS-based bioanalyses, herein we will briefly survey the ones with reliable clinical perspectives in diagnosis.
Below, we overview the use of SERS substrates in bioanalysis and disease diagnosis; readers will find details of recent progress in SERS-based bioanalyte detection in each particular section of this review. First, this review sets out the basic mechanisms that empower SERS to the point of trace-level detection. Next, it highlights the strong advantages and the limitations of SERS, covering label-free detection, direct identification of molecules in situ, standoff detection, etc. These are key aspects for successful SERS-based approaches as part of infection diagnosis and clinical pathways. Finally, this review examines the promising future research directions for SERS as a spectroscopic analytical method in infection diagnosis and targeted treatment strategies. This review article is intended as a useful tool for performing and developing bioanalytical SERS assays and for employing the full potential of SERS-based in vivo applications in bioanalysis and diagnosis.
2. SERS substrates
The illumination of metallic nanostructures with the monochromatic light of a laser leads to an excitation of the collective oscillations of surface conduction electrons, known as surface plasmons. Surface plasmons could be considered as vibrating electric dipoles that create a strong electric field in their close proximity. In metallic nanoparticles, the surface electrons are subjected to the competing forces of the electric field of light and of the restoring forces from positively charged nuclei. A simple harmonic motion results with an intrinsic resonant frequency – the localized surface plasmon resonance (LSPR). Excitation of the LSPR results in enhancement of the vibrational scattering signal from those analyte molecules that are found in the first tens of nanometers from the metal surface. Consequently, SERS is extremely dependent on the substrate. Substrates consisting of so-called plasmonic nanoantennas present resonance frequencies in the Vis-NIR region that can be tuned by varying the size and shape of the nanostructures.24 For instance, for gold nanospheres, the LSPR wavelengths are between 520 and 530 nm for Au NP radii ranging from 8 to 20 nm.25The utmost specific criteria that SERS-active substrates need to fulfil are the predictable and reproducible SERS enhancement, the LSPR availability for optimization at a desirable wavelength,26 a large area of uniformity and performance.27 For instance, a helpful technique to tune the LSPR position across the visible and near-infrared regions while recording the SERS signal at a fixed Raman excitation wavelength is plasmon-sampled surface-enhanced Raman excitation spectroscopy (PS-SERES).28 With this technique, the SERS enhancement factor (EF) is investigated both as a function of the LSPR spectral location (ranging from 350 nm to 1900 nm) and as a function of the input laser wavelength (between 633 nm and 1064 nm). By tuning the LSPR in this manner, one of the highest EFs was measured.
The fundamental metric for the efficiency of the SERS effect is the EF, ranging from 102–1014 and quantifying the enhancement in SERS signal intensity (counts per s per mW) per molecule. Determining the surface coverage of a Raman probe and calculating the enhancement factor of the SERS-active substrate is necessary, but also not very clearly established. The extent of SERS applications by using various plasmonic materials makes this estimation less coherent. In some cases, the EF is measured by calculating the number of molecules adsorbed on a metallic surface area,29 the concentration of the analyte, scattering volumes, and laser powers at each wavelength30,31 or the intensity of a selected spectral band divided by that of a standard, such as ethanol (which does not present a significant SERS signal).32,33 However, there are several factors, including packing density, surface roughness and area, that make it challenging to determine the SERS EF from the number of molecules. Hence, the number of adsorbed molecules can be determined from the best estimated packing density value for a known order of surface probe molecules. One needs to also consider that the number of molecules involved in the SERS signal is practically derived from the absolute signal intensity, but only approximated due to the extreme statistical distributions.
Le Ru et al.34 evaluated the EF in their measurements. The SERS substrate enhancement factor is commonly defined as follows:
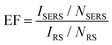 | (1) |
While the single molecule is considered the ultimate detection limit, in real-life applications, sample uptake is minimal, ranging from sub μg to ppb levels. Due to the consistent contribution via electromagnetic enhancement, a strong SERS signal should be provided for individual molecules. If labelling is an option, “SERS labels” can give real “overvalues” in comparison with fluorescence tags. The ultra-high specificity of SERS makes it almost impossible to observe overlapping between different labels even if they have a similar structure. Their affinity to the metallic surface used as a SERS-active substrate and the continuous competition between species to adsorb on this surface need to be taken into account. The excitation wavelength used also “selects” only the particular species by resonance conditions. For instance, resonant Raman or SERS spectroscopy was also used for studying many “intrinsically colored” biomolecules such as chlorophylls,35 haem-containing proteins,36 carotenoids,37 rhodopsin,38 and flavin nucleotides,39 and with the aid of robust chemometric tools, their accurate detection and discrimination in living cells was successfully reported.40
There are numerous opportunities to use SERS in real multiplex detection for screening biological processes, real-time mapping of living cells, etc. Additionally, the SERS approach is insensitive to photobleaching and self-quenching of the tag due to the non-resonant fundamental SERS enhancement mechanism. In the following, we present particular preparation techniques for obtaining optimal platforms for SERS-based detection in clinical premises.
2.1. Rational design of high-performance SERS substrates
The bottom-up and top-down methods for preparing tunable, highly sensitive and rapid SERS-active substrates of various thickness and efficiency are detailed in this sub-section.The drop-coating deposition Raman (DCDR) method consists in applying a 1–5 μL droplet of mixed NPs and analyte, which is afterwards mapped or just irradiated continuously until the droplet has dried. This method allows time- and/or spatially-resolved SERS mapping with the advantage that it yields high SERS intensities due to the formation of a three-dimensional (3D) hotspot matrix during the drying process.41 DCDR is also employed to assess the limits of spectrum reproducibility, and possible photothermal degradation under laser excitation, and to attain robust, reagent-free and sensitive analyte detection, especially in the case of protein analysis at low concentrations.42 The technique provides a sensitive, rapid and reproducible method to achieve Raman spectra from low protein concentration solutions.43 For instance, combining SERS analysis of serum with standard testing appears promising for a more accurate diagnostic rate of prostate cancer.44 Moreover, multiplex, reagent-free assays were devised for glycated albumin level determination by combining Raman spectroscopy with multivariate classification techniques45 or for SERS-based testing for microalbuminuria in urine samples.46
Another rapid and highly sensitive approach is the in situ synthesis of Ag colloids47–49 with a potential to develop point-of-care (PoC) diagnostics. This biosensing method is emerging as an alternative to the predominant label-free SERS-based approach for the identification of bacteria, which involves simply mixing presynthesized Ag NPs with the bacterial biomass. In this method, probing the bacterial fingerprints is difficult due to the low spectral reproducibility. The in situ approach allows highly intense SERS spectra by providing a direct contact between Ag NPs and the bacterial membrane. The bacteria and NPs are brought together by the strong electrostatic attraction between the negatively charged bacteria and the positively charged Ag cations. This induced adhesion to the cell wall was also reported most recently for cyanobacteria.50 The cell wall intrinsic SERS patterns that are achieved by label-free detection are employed for the simultaneous detection of different pathogens.48,51,52 However, the sensitivity and validity of this label-free (direct) SERS-based approach is dependent on the SERS substrate, and this technique can be challenging in complex conditions, such as biofluid samples. Moreover, the spectra from distinct bacterial species can be very similar. Consequently, suitable discrimination models and chemometrics tools need to be implemented.
The control achieved by using bottom-up methods is possible by tuning the size and shape, and by employing various compositions for the designed nanostructures and clustering strategies in order to generate high local electromagnetic field enhancement or “hotspots”.53 Silver and gold are the most used in chemical synthesis approaches, providing a plethora of geometries: spheres,54–57 nanocubes,58 tetrahedra, pyramids, nanoprisms,59 nanorods, nanoflowers,60 popcorn-shaped,61 ribbons, nanoshells, nanosprings,62 nanobelts,63 nanostars,64,65 nanocrescent shapes, dumbbells,66 urchin,67etc. (Fig. 3).
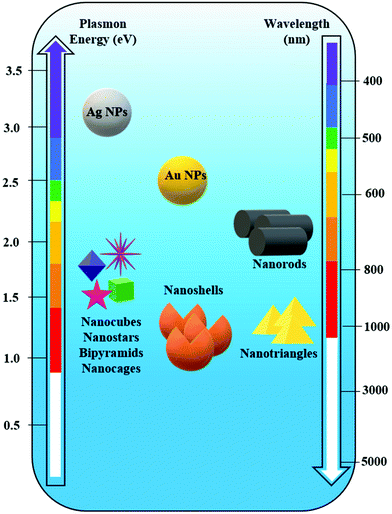 | ||
| Fig. 3 Spectral dependence of the plasmon resonance of differently shaped silver (Ag) and gold (Au) nanoparticles (NPs). | ||
Recent progress in one-pot synthetic procedures for the preparation of anisotropic nanoparticles is significant despite the high sensitivity to chemical changes and the difficult control of size and morphology (flocculation, agglomeration or clustering effects). Sometimes, in order to prepare large particles by a seed-mediated process, a step-by-step growth would be recommended, to avoid nucleation. Particle characterization is essential to guarantee that samples from different batches have identical physical and chemical properties. Systematic batch testing by various characterization methods needs to be employed.68
The influence of NP aggregation on SERS signal intensity is well known, and according to previous studies it was observed that chemically induced aggregation,69,70 agglomeration and oxidative etching,71 iodide modification46,72 and aggregation by multiple NP monolayer deposition cause intensity changes.3,73 Thus, it can be observed that the average SERS intensity increases with the increase in particle aggregation. By contrast, Wustholz et al.74 achieved a manipulation of NP aggregation by employing field flow fractionation and they revealed that it is not essential that SERS enhancement increases with the cluster size. In their findings, it was observed that SERS intensity provided by NP dimers was comparable to that of NP heptamers, the enhancement being more related to the size of the interparticle gap. It is widely acknowledged that interstices at the junction between adjacent NPs act as “hotspots”57,75,76 (initially called “hotsites”53) which provide electromagnetic enhancement (EM) resulting in a 108 times larger SERS signal in contrast to normal Raman scattering (Fig. 4A–C).77 Moreover, Shaw et al.68 have reported a statistical approach that allows the sampling thousands of NP aggregates, to provide a comprehensive overview of the relationship between SERS intensity and NP cluster size (Fig. 4D). By combining images of probe clusters (obtained by transmission electron microscopy (TEM), atomic force microscopy (AFM), and scanning electron microscopy (SEM)) with SERS analysis from exactly the same regions of the sample, this approach helps to determine the SERS intensity and cluster size relationship. By applying this statistical approach, information on the changes in SERS intensity for the probes is obtained. The probes are specially prepared for target-specific SERS applications that also provide biological cell imaging. The study of the cluster SERS intensities could be directly applied in vitro.
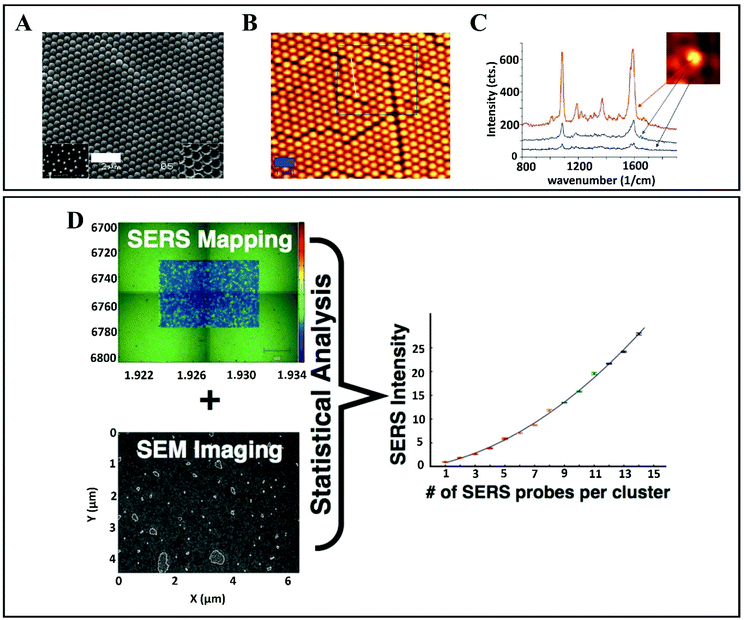 | ||
| Fig. 4 (A) Scanning electron microscopy (SEM) and (B) atomic force microscopy (AFM) images of gold films over nanosphere (AuFoN) substrate. (C) SERS spectra measured at three different regions: red (hot-spots), blue (adjacent to half-shells) and black (gold half-shells). Reproduced with permission.57 Copyright 2010, American Chemical Society. (D) SERS map and SEM image of the surface of gold slide after incubating it with SERS probe. Statistical correlation between SERS probes per cluster and mapped SERS intensity which enables it to sample the thousands of NPs aggregates. Reproduced with permission.68 Copyright 2013, American Chemical Society. | ||
Silver-based substrates are known to offer high EFs due to the particular optical properties of silver, which restrict interband transition and thereby favour plasmon resonance.66 The fine manipulation for tuning the shape, size and assemblage of silver NPs (Ag NPs) is achieved by either chemical interactions or physical forces. Chemical interactions control the formation of dimers/trimers using linking molecules or macromolecules (graphene, DNA, etc.). The constructive application of Ag nanostructures in biosensing with fit-to-purpose clinical perspectives was recently reviewed by Tan et al.78 Generally, modified Ag NPs are recommended as biosensing platforms with improved performance in life-threatening diseases, such as cancer, HIV and viral/bacterial infections. Subsequently, silver (and gold) nanomaterials serve as prime candidates in increasing the SERS signal and in improving the detection sensitivity. They offer rapid, efficient, cost-effective and robust SERS platforms for detecting trace levels of biomolecules and pathogens, at single-cell level, due to their unique chemical, physical and optical properties. For instance, core–shell approaches such as Au@Ag nanoparticles serve as excellent SERS substrates, since significantly higher EFs are expected for silver as compared with gold.79–81 When using gold nanomaterials for instance, one drawback might be the high photothermal conversion efficiency provided under resonance excitation, which can be destructive for biological samples. Thus, the rational choice of the shape and composition, and tuning of the excitation spectral window for a minimal heating and photodegradation are the key elements in using nanostructured probes in biosensing. Also, bare/nascent Au NPs are relatively unstable under physiological conditions due to the formation of large aggregates that hinder their use in vivo.
However, tailored Au NPs can be manipulated into multifunctional, optical sensors: they are either used for their colour change and/or their SERS properties in combination with biocompatible polymers, oligonucleotides, proteins, macromolecules, antibodies etc.82,83 There is frequently a covalent bonding between the biomolecules and ligands onto the NPs’ surface by using oligoethylene (OEG) or polyethylene glycol (PEG),84,85 which are stable spacers. More importantly, they assure specific adsorption of only the wanted biomolecules or are meant to facilitate in vitro SERS-based cellular testing and to prevent NP aggregation.86
Since 1984, Vo-Dinh's team has been designing Nanowave-based SERS substrates with very high efficiency.88–91 Their fast, cost-effective, sensitive and label-free approach for the detection of DNA with applications in clinical diagnosis is represented by an unique biosensor based on a molecular sentinel (MS)88 that is dispensed on the surface of a plasmonic Nanowave chip (MFON). Its sensitivity is dependent on the decrease in SERS intensity when there is a physical separation of the Raman label tagged at one end of the MS from the MFON's surface due to DNA hybridization. A schematic illustration of the detection mechanism is shown in Fig. 5A. Firstly, the surface of the MFON is functionalized with an MS. A hairpin loop structure is formed after the hybridization by the complementary arms into 6 basepair stem sequences. The Cy3 Raman label is attached at the 3′-end of the MS hairpin probe. At the 5′-end, there is an alkyl thiol substituent that favours conjugation between the MS probe and the MFON's gold surface. In the presence of a complementary target DNA sequence, the MS's stem loop opens and the SERS signal is quenched due to separation of the Cy3 dye from the Au surface (Fig. 5B). However, there is a strong SERS signal in the absence of a target DNA sequence, because the MS's stem loop remains in a closed state, as there is a close proximity between the Cy3 dye and the Au surface (Fig. 5B). Such direct (label-free) approaches are more and more present in clinical diagnosis research based on spectroscopic techniques.
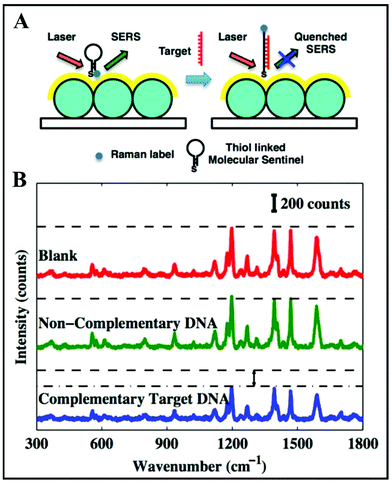 | ||
| Fig. 5 (A) Schematic of the detection scheme of label-free complementary target DNA. (B) SERS spectra obtained from blank (red), non-complementary DNA sample (green) and complementary target DNA sample (blue). Reproduced with permission.88 Copyright 2013, American Chemical Society. | ||
A modern approach consisting in designing wearable, reusable textiles with plasmonic properties has been developed.92 Practically, creating micropatch arrays of Au NP aggregates on nanoporous semipermeable transparent membranes that are further nanoimprinted on a stretchable fabric by UV-resist micro/nanofabrication techniques is demonstrated (Fig. 6A and B). The textiles are washable and reusable by simply using detergents. These membranes were initially designed as wearable biochemical sensing devices, for injury or body fluid monitoring. Yet, they were also successfully tested on fresh meat. The sensing potential was demonstrated by detecting the highly fluorescent dye rhodamine 6G (R6G) at different concentrations from 10−3–10−7 M on the meat surface as an in situ SERS monitoring approach. As shown in Fig. 6C, there is an additional peak at 727 cm−1 revealed by “SERS membrane + 10−3 M R6G + meat” as compared with the SERS spectra of “SERS membrane + meat” and “SERS membrane + 10−3 M R6G” that indicates the presence of adenine molecules on the meat. Bacteria release such adenine as a metabolite.93,94 Such bacteria can be present on raw poultry, and therefore it can be inferred that the Raman peak at 727 cm−1 is due to adenine-containing metabolites secreted by these bacteria. Hence, the SERS membrane can be utilized to perform on-site rapid detection of meat spoilage. The SERS performance of the designed textiles was proved along with the versatility of top-down nanofabrication approaches, using template-assisted self-assembly and micro/nanoimprinting.
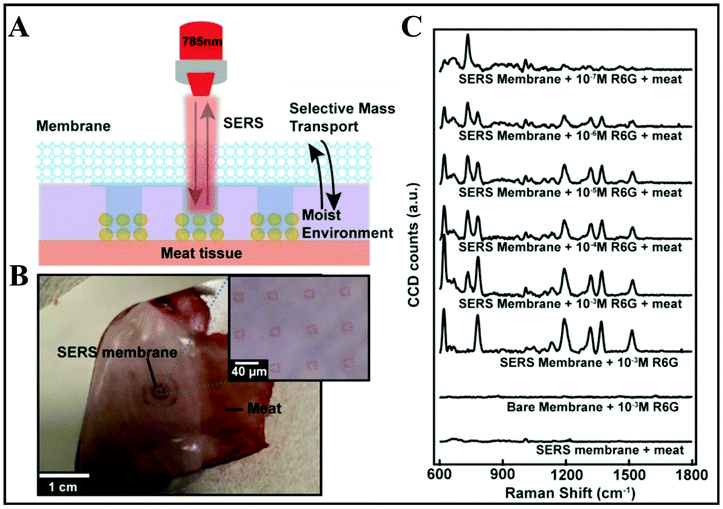 | ||
| Fig. 6 In situ SERS analyses of meat surface by using SERS membranes. (A) Schematic of the experimental procedure. (B) Top-down camera picture and bright-field micrograph of the SERS membrane on the meat surface. (C) SERS spectra obtained from SERS membrane on meat, the bare membrane with 10−3 M R6G, SERS membrane with 10−3 M rhodamine 6G (R6G), and SERS membrane on meat with R6G at different concentrations of 10−7, 10−6, 10−5, 10−4 and 10−3 M. Reproduced with permission.92 Copyright 2020, American Chemical Society. | ||
So far, SERS reproducibility has been improved by developing more uniform, SERS-active substrates through novel top-down nanofabrication techniques but this approach has high costs for scale-up. Alternatives come from the idea of improving point-to-point and batch-to-batch variability caused by the heterogeneity of the SERS substrates or hotspot-related variations. Isotope-edited internal standards (IEIS) were tested as candidates for SERS quantitation even under dynamic SERS measurements when hotspot density can change in time.95,96 IEIS are ideal due to their identical Raman cross-section and affinity for the plasmonic surface as of their isotope analogues. So, the quantification of the analyte concentration is related to the ratio of the Raman band intensities of both isotope analogues.
2.2. Plasmonic hotspot engineering
To attain reproducible and reliable SERS measurements in high-performance SERS devices, it is necessary to (nano)engineer plasmonic hotspots, for instance based on in-plane and out-of-plane vertically stacked plasmonic nanogaps.Zhang et al. have reported a synthesis of 3D hierarchically porous metamaterial by a dual-templating technique in which non-ionic surfactant octaethylene glycol monohexadecyl ether (C16EO8) is used for a lyotropic liquid crystal (LLC) and reverse porous poly(methyl methacrylate) (PMMA) is used for a hard template.107 The steps involved in the preparation of porous gold metamaterials are shown in Fig. 7A. Firstly, a negative PMMA template is prepared from porous anodic aluminum oxide (AAO) template. Afterwards, the LLC precursor solution is injected into the PMMA template for the formation of LLC within nanovoids of the PMMA template. Then it is followed by gold electrodeposition, which results in PMMA template removal. This 3D plasmonic metamaterial-based SERS substrate (i.e. hierarchically ordered porous gold membranes) comprises uniformly scattered mesopores and closely packed nanohole channel arrays.107 These nanoholes have LSPs that allow the harvesting of incident light efficiently, producing strong EM enhancement within close proximity to the nanohole edges on the Au substrate. There is cascaded multiscale EM field enhancement observed by the plasmon modes, due to the incorporation of arranged mesoporous nanostructures into the nanohole arrays. As a result, strong Raman-active sites are generated on the entire substrate. In addition, efficient binding sites are also provided by mesopores to capture the analyte molecules at hotspots. These are the advantages that make 3D plasmonic metamaterial-based SERS substrate an efficient one.
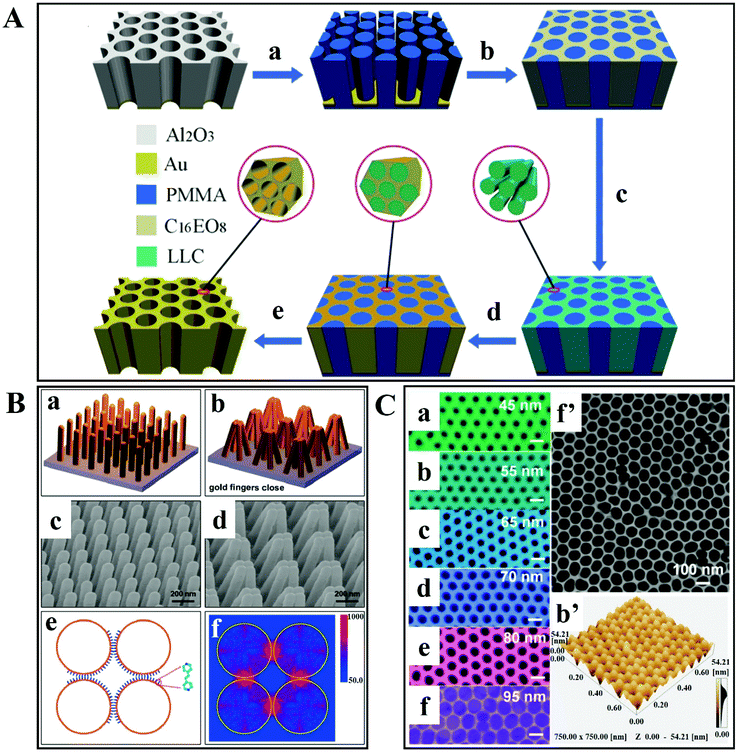 | ||
| Fig. 7 (A) Schematic illustration of the preparation of 3D hierarchically porous metamaterials. (a) Synthesis of negative poly(methyl methacrylate) (PMMA) from porous anodic aluminum oxide (AAO) template; (b) injecting the lyotropic liquid-crystal (LLC) precursor solution into the PMMA template; (c) formation of LLC within the PMMA template; (d) gold electrodeposition; and (e) PMMA template removal. Reproduced with permission.107 Copyright 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (B) Gold-coated nanofingers. (a and b) Schematic of gold nanofinger closure due to capillary force. (c and d) Scanning electron micrographs of open and closed gold nanofingers. (e) Schematic representation of the molecule-trapping within the finger nanogaps. (f) Electric field intensity |E(r)|2 distribution at 750 nm for the gold spheres having a radius of 68 nm. Reproduced with permission.108 Copyright 2010, American Chemical Society. (C) SEM images of ultrathin alumina mask of different pore diameters: (a) 45 nm, (b) 55 nm, (c) 65 nm, (d) 70 nm, (e) 80 nm and (f) 95 nm. (f′) SEM image of a large-area ultrathin alumina mask with a pore diameter of 95 nm; and (b′) the AFM image is in good agreement with part (b). Scale bars: 100 nm. Reproduced with permission.109 Copyright 2015, American Chemical Society. | ||
A very interesting mechanism for capturing analyte molecules in a solution was reported by Hu et al. (Fig. 7Ba).108 It consists of a molecular trap structure of free-standing polymer nanofingers created by nanoimprint lithography. These nanofingers are coated with Au and they can trap the analyte molecule using the microcapillary force upon liquid exposure. Hotspots are generated at the same time on the tips of the Au-coated flexible polymer fingers, which enables SERS detection and analyses. An SEM image of an Au-coated flexible nanofinger array is shown in Fig. 7Bc. After exposing the array of Au-coated nanofingers to the analyte solution and then air-drying, the nanofingers close in groups of four as represented in the schematic illustration in Fig. 7Bb and the SEM image in Fig. 7Bd. After the liquid evaporates from the array, the nanofingers are pulled towards each other and the analyte molecule is trapped between the fingertips. To model the Au fingertips, discrete dipole approximation (DDA) was utilized (Fig. 7Be). The electric field map plan view of the Au gold spheres for 750 nm unpolarized incident radiation is depicted in Fig. 7Bf. It is also observed that the strongest field is concentrated among the four gaps of the sphere. In this study, the SERS-based sensitive detection of molecules is ensured by the capability of controlling the self-limiting gap size between the fingertips.108
In order to confine electromagnetic energy within subwavelength dimensions, there are gaps,110,111 holes,112,113 tips,114 slits115 and metal particles116 that can be designed. The confinement with the greatest degree can be achieved in a gap of nanometre scale between two metal surfaces.116 Electron beam lithography,117 electromigration118 and scanning probe119 techniques can be useful in creating point-like nanometric junctions but they are not practical for the preparation of devices with Ångstrom-scale dimensions over a large area.
As an example of an alternative, Chen et al. have devised a technique based on atomic layer lithography in a combination of atomic layer deposition (ALD) with ‘plug-and-peel’ metal patterning to generate nanometre-scale gaps over millimetre-scale contours that allow the resonantly enhanced transmission of terahertz waves across opaque metal films.120 Upon directing terahertz waves towards a 1 nm gap, field enhancement factors of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 can be observed.
000 can be observed.
In another example, Fu et al. have reported an ultrathin alumina mask (UTAM). This surface-pattern-based, cost-effective technique results in interparticle gaps of 5 nm.109 Ag was used and the Ag deposition, thickness and evaporation rate are directly proportional to the shape, density and size of the Ag NPs. In order to achieve well-aligned and uniform Ag NPs, the nominal layer thickness was 50 nm and the evaporation rate was 0.4 nm s−1. Fig. 7C shows the scanning electron micrographs of UTAMs with different pore diameters. By controlling the UTAMs’ structural parameters, the Ag NP arrays are arranged in a highly ordered manner over a large area, as shown in the SEM images. It is also observed that there is a decrease in interpore distances from 55 to 5 nm on increasing the UTAM pore size from 45 to 95 nm (Fig. 7Ca–f). The high uniformity and regularity over a large area of UTAMs with a pore diameter of 55 and 95 nm is shown in Fig. 7Cb′ and f′. The value of relative standard deviation (RSD) measured from 10 random spots is about 2%, which indicates highly reproducible signals. Furthermore, finite-difference time-domain simulations reveal that narrow gaps are producing enhanced electric fields. Such a closely packed array of Ag NPs in the UTAM with a lower RSD value and high density of electric field enhancement enables uniform SERS detection with high sensitivity and excellent reproducibility.
Song et al. have reported a preparation of scalable high-performance SERS substrates based on multistack vertically oriented nanogap hotspots in nanolaminated plasmonic crystals, that can be synthesized by a top-down preparation process (Fig. 8).121 With the help of UV-curable polyurethane (PU) polymer resist, the polymer nanopillar arrays are replicated on polyester (PET) films by using a nanostructured polydimethylsiloxane (PDMS) stamp. Afterwards, Au and SiO2 layers are alternatively deposited by using electron-beam deposition, in order to prepare metal–insulator–metal nanostructures having a multistack vertically oriented plasmonic nanogap on each nanopillar. Wet chemical etching is applied to reveal the embedded nanogap hotspots with buffered oxide etchant (BOE) 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. This study constitutes a promising example of an out-of-plane plasmonic array for a high-performance SERS substrate for biochemical analysis.
1. This study constitutes a promising example of an out-of-plane plasmonic array for a high-performance SERS substrate for biochemical analysis.
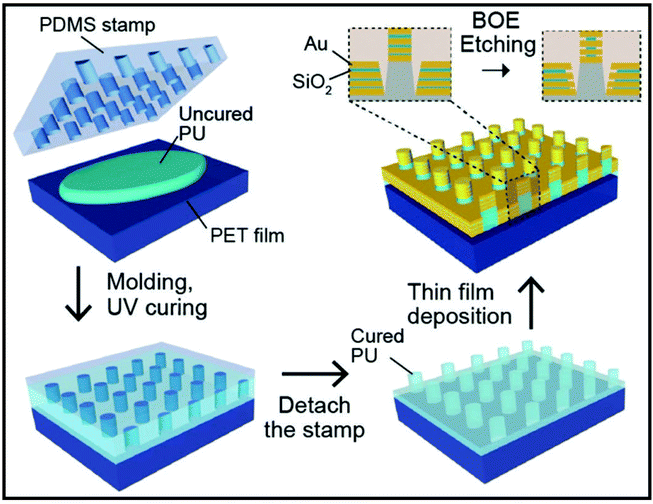 | ||
| Fig. 8 Schematic of the preparation of nanolaminated SERS substrate with three insulator layers. Reproduced with permission.121 Copyright 2019, WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
Furthermore, Nam et al. have reported a multi-layered metal–insulator–metal plasmonic nanostructure-based nanolaminated SERS substrate with uniform hotspots and a significant EF (>107).122 In the case of conventional SERS substrate, the EF is dependent on the background refractive index (RI). In the nanolaminated SERS substrate, there are uniform arrays of vertically oriented nanogap hotspots that are insensitive to variations in background RI. Experimental and numerical studies suggested that broadband multiresonant optical properties of the nanolaminated plasmonic nanostructures are behind the RI-insensitive SERS response. Such a high-performance SERS substrate reveals molecular profiling and classification of breast normal cells and living breast cancer cells with a 96% high prediction accuracy. It could also help in investigating the spatiotemporal biochemical dynamics in cellular networks.
2.3. Internal-standard-based SERS calibration techniques
In order to improve the quantitative SERS and multivariate analysis for biomedical applications, there are two main calibrations that are of interest: the molecular Raman tag-based internal standard and the background-scattering-based internal standard.As a solution to work around this issue, a Sweden research group proposed three internal standard solutions (4-mercaptobenzonitrile – MBN, 3-mercaptopropionitrile – MPN and 4-cyano-N-(2-mercaptoethyl)benzamide – CMEB respectively)123 for the detection of rhodamine R6G using gold colloids. The strong distance-dependence of the enhancement effect was exploited by using the aforementioned alkanethiolate as self-assembled monolayers (SAM) attached to Au NPs.123 The three cyano-containing compounds were selected due to their chemical stability in monolayers where they are firmly attached to the metallic surface through thiol bonds and for a particular silent band assigned to the CN vibration at 2300 cm−1. The internal standards are meant to uniformly cover the metal surface as a monolayer and to prevent chemisorption of the analyte, which could negate their function. For calibration, the signal of MBN, as internal standard, and the signal of R6G, as analyte, were monitored at different concentrations and different proportions between them. By using 4 partial least squares (PLS) components, 2 for variations in the spectral baseline and 2 for the magnitude and proportion between the analyte and the internal standard, a quantitative calibration model was developed.123
For quantitative SERS measurements down to single-molecule level, alkanethiolates were also used as ligands for close-packed Ag NPs superlattices with high uniformity and a large enhancement factor.124 In this study, the analyte molecules, crystal violet (CV), were embedded in an ultrathin spin-on-glass (SOG) “hot zone” of only 5 nm prepared as a planar uniform layer above the Ag NP film with high SERS performance (Fig. 9A).124 The signal from the thiolate ligands was used as the internal calibration standard for the quantitative SERS measurement of CV molecules areal density. The calibration of the CV SERS intensity (1177 cm−1 marker band selected) is performed as a ratio of the SERS intensity from CV and alkanethiolate. This is possible because the SERS intensity of the CV molecules changes proportionally with their areal density whereas the SERS intensity of alkanethiolates remains at the same level.
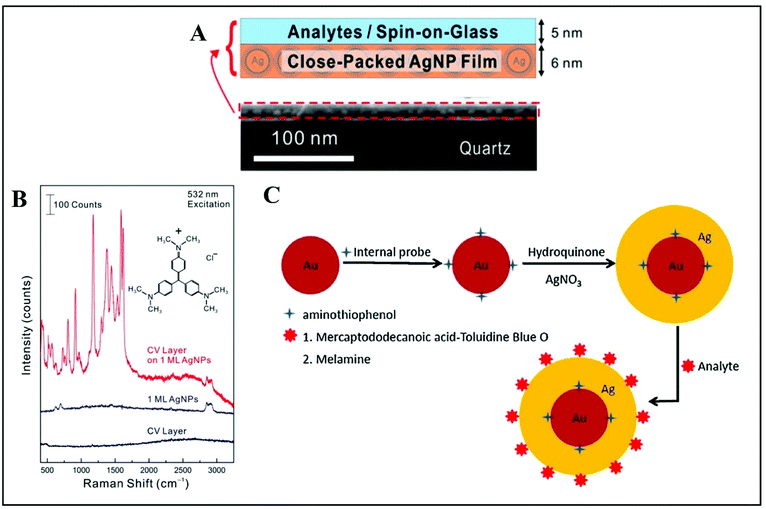 | ||
| Fig. 9 (A) The analyte is a spin-coated on a glass (SOG) layer of crystal violet (CV) molecules. The SERS substrate is a closely packed Ag NP film. The CV/SOG layer and Ag NP film thicknesses are about 5 and 6 nm respectively. The SEM image shows both films on top of a quartz substrate. (B) Raman spectra recorded from a CV/SOG layer on a Ag NP film, a bare Ag NP film and a bare CV layer, respectively. Reproduced with permission.124 Copyright 2015, American Chemical Society. (C) Schematic representation of quantitative SERS measurements by utilizing internal reference embedded Au-core/Ag-shell NPs: adsorption of the probe molecule serving as internal reference (4-ATP) onto the Au surface, followed by deposition of a Ag layer over the top surface; the approach is used either for Toluidine Blue O (TBO) detection through a linker molecule, 12-mercaptododecanoic acid (MDA), and or for direct melamine detection, without the linker molecule. Reproduced with permission.126 Copyright 2015, Elsevier B.V. | ||
The advantage of employing thiolate ligands on the Ag NP surface is that the collective plasmon modes can be tuned by exploiting the plasmonic coupling effect. Practically, by varying the carbon-chain length of the ligand, an interparticle space is defined which hugely influences the near-field coupling strength between adjacent Ag NPs.125 Finite-difference time-domain (FDTD) simulations were performed to estimate the field distributions in the depth of the “hot zone”. The electric field intensity distributions in different vertical planes on top of the Ag NP film were revealed to be highly uniform with a standard deviation of field intensity at the hot zone of only 10% of the peak value. It is noteworthy to underline the fact that this approach was developed for a specific wavelength of excitation that matches the plasmonic resonance peak, namely the 532 nm laser line; otherwise the Raman enhancement is not observed (Fig. 9B). Tuning the plasmonic properties would imply preparing another Ag NP film with a different ligand than octodecanethiolate.124 The strong Raman enhancement obtained for CV suggests a single-molecule yield and potential implementation in microfluidics for analytes in liquid solutions.124
An original way to embed internal standards by employing core–shell Ag or Au-based NPs is addressed by Zhou et al.126 and Shen et al.,127 for instance. Silver-based quantitative SERS measurement with embedded 4-aminothiophenol (ATP) as internal reference was implemented for detecting melamine in milk.126 Melamine is an omnipresent adulterant mainly in dairy products that are tested for their protein content.128,129 Its detection by using Au-core/Ag-shell NPs was demonstrated by Zhou and co-authors successfully and after optimization, and their simple and rational approach could be translated to a wide range of other analytes.126 The proof-of-principle is based on adsorption of the probe molecule serving as internal reference (4-ATP) onto the Au surface, covered afterwards by a silver layer. The approach can be used with a linker molecule, as demonstrated by the authors when detecting Toluidine Blue O (TBO) through 12-mercaptododecanoic acid (MDA) or without this step, by directly mixing the core–shell NPs with the target analyte (melamine, for instance) (Fig. 9C). Thus, the internal reference is embedded between the Au core and Ag shell and the target molecule is adsorbed onto the outer Ag shell. Subsequently, the detection is only a question of the adsorption ratio onto the surface for the analyte and a careful selection of the spectral bands monitored. The peak for the analyte and the one monitored for 4-ATP should be distinct. Limits of detection reported are considered useful in real-life applications. The versatility and high performance of this approach recommends it for further implementation by using various analytes.
A novel type of internal standard approach for reliable quantitative SERS analysis is proposed by Shen et al.127 through exploiting the advantages of core–shell nanostructures. The rational selection for the Raman tag (4-mercaptopyridine – Mpy) and the cysteamine as a framework for Ag shell formation is guided by the applicability of this synthesis method. A strong signal is recorded from the Raman tag, independently of the outer environment, and the SERS signal of the target molecule (1,4-phenylene diisocyanide – PDI) is monitored in real time for nM concentrations. Mpy and PDI have different spectral windows for their strong bands and can be simultaneously monitored. Thus, the quantitative SERS analysis was performed by normalizing the PDI signal (2184 cm−1 selected band) to that of Mpy (by using the 1095 cm−1 band) at different concentrations. A strong point was to demonstrate that analytes with less affinity for the metallic surface can also be effectively captured for detection, by using a forbidden dye (basic red 9) from the textile industry. Thus, a highly sensitive, label-free, versatile and reliable method for direct quantitative SERS analysis was developed.
For instance, Vikesland et al. proposed to calibrate the SERS signal from the analytes with the intensity of the surface-plasmon-enhanced Rayleigh band IRayleigh originating in the amplified spontaneous emission (ASE) of the laser.130 This IRayleigh appears quite suitable for reference purposes; it was tested in six SERS-active substrates and was found to be a reliable quantitative tool for predicting the efficiency and reproducibility of the SERS substrates. IRayleigh reflects the “hot spot” efficiency within the laser excitation volume irrespective of the physical particularities of the nanostructures used in colloidal or solid phase. As defined by the equation below, IRayleigh is in a SERS experiment proportional to IRaman since only NA is not constant:
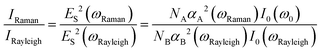 | (2) |
Vikesland's group developed and improved this coefficient of variation for a SERS substrate.131 The normalization of hot spots is independent of the incident laser intensity and is based on a localized intrinsic internal standard (υe) that scales across all plasmon-enhanced electromagnetic fields within the substrate tested. The surface-enhanced elastic scattering signal (IRayleigh) originates from the amplified spontaneous emission (ASE) and has a characteristic pseudo-band at low wavenumbers due to ASE interaction with an edge filter (Fig. 10A). An important aspect reported by the authors is that the point-to-point variability of the tested substrates after the SERS signal normalization technique was applied is hugely reduced to less than 10%. It was demonstrated that the intensity of the low wavenumbers pseudo-band (υe) scales linearly with the integrated “hot spot” signal strength, so υe can be used for calibration in order to obtain a minimized signal variation for the SERS substrate. The only disadvantage that Raman users might find for this quantitative SERS hotspot normalization technique is the need for bandpass/edge filters introduced in the measurement setup. This is not feasible for many portable, compact or handheld Raman systems (Fig. 10A).
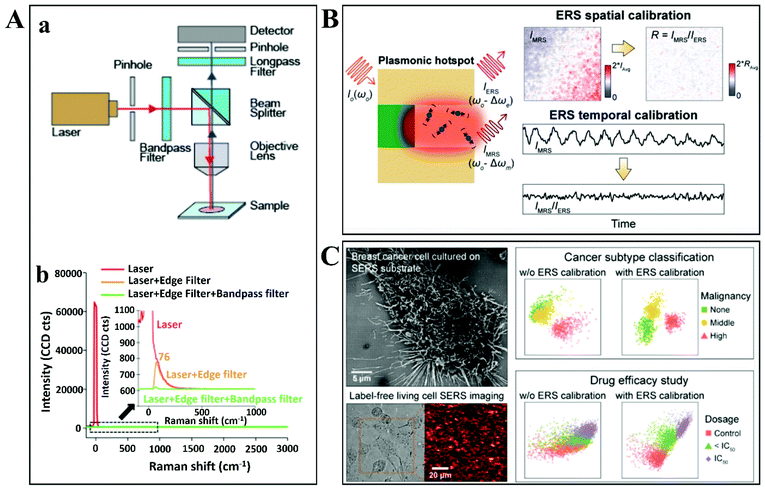 | ||
| Fig. 10 (A) An experimental setup design for simultaneous collection and comparison of inelastic Raman and elastic Rayleigh scattering signals from the focused spot on the nanostructured plasmonic substrate. (a) The design and configuration used during backscattering confocal Raman microspectroscopy measurements. (b) The laser emission spectra recorded in transmission mode with or without an edge filter and bandpass filter. Reproduced with permission.131 Copyright 2018, American Chemical Society. (B) Illustration of plasmonic enhancement of ERS and MRS signals at hot spots along with representative 2D Raman images for IMRS before and after ERS spatial and temporal calibration. Reproduced with permission.132 Copyright 2020, American Chemical Society. (C) Top view of an SEM image showing the highly malignant breast cancer cells cultured on the specially designed nanolaminated SERS substrates, along with bright-field and 2D Raman mapping images of the same living cells. Representative PLS-DA scatter plots are shown alongside for subtype classification and drug efficacy improvements due to ERS-based SERS calibration. Reproduced with permission.133 Copyright 2021, American Chemical Society. | ||
Most recently, nanolaminated SERS substrates were proposed as generalized metallo-dielectric plasmonic systems that could support both localized and delocalized plasmonic modes. The plasmonic electronic Raman scattering (ERS) signals from the metal were used as an internal standard for spatial and temporal calibration. By using a long-pass filter to block the laserline (Rayleigh scattering), plasmonic ERS signals originating from the surface-plasmon-enhanced inelastic light scattering of sp-band electrons in the metal are visible as an ERS pseudopeak (87 cm−1) in the filtered SERS background signal. Thus, the molecular Raman scattering (MRS) signal of the analyte can be calibrated by using the ERS signal as internal standard, at the same plasmonic hot spots. The ratio between MRS and ERS signals from the same hot spots is approximated by the authors as follows:
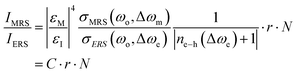 | (3) |
Quantitative SERS analysis was performed by ERS-calibrating the MRS signals for benzenethiol (BZT) and rhodamine (R6G) molecules (Fig. 10B).132 ERS-calibrated Raman signals were verified and found to be insensitive to variations of local field intensities from different hot spots by spatial correlation. 2D mapping of surface-immobilized test molecules showed significant improvement of the SERS quantitative analysis by removing the effects of local field variations among hot spots. The ERS internal standard was evaluated for temporal SERS calibration also, by monitoring the intensities ratio (MRS signal vs. ERS signal) in response to static and dynamic laser excitation fluctuations.
As a direct application of this concept, ERS-based SERS calibration was used for label-free SERS analysis of living cells for monitoring drug response and degrees of malignancy (Fig. 10C).133 Nanolaminated metal–insulator–metal (MIM) nanostructures were used for SERS profiling and 2D Raman imaging for breast normal and cancer cells. After ERS-calibration of the SERS spectra and by using multivariate analysis (PLS-DA), three degrees of malignancy were discriminated: non-malignant, moderately and highly malignant. Furthermore, the effects of ERS-calibration on statistical bioanalysis performance in drug efficiency were investigated. IC50 values for PTX in three different dosages were determined by 2D SERS mapping measurements of living cells under treatment. ERS-calibration was able to enhance the supervised learning classification of treated/untreated cells but is not definitive due to significant overlapping between groups with an order of magnitude difference in dosages.
3. SERS-based pathogen detection methods
Pathogen detection is one of the major human health problems. Fast, ultrasensitive and efficient tools are needed to accurately assess the spreading of infections and to guide treatment.Pathogenic bacterial infections constantly give rise to serious challenges in diagnostics, despite the availability of modern molecular techniques. Regardless of the infection type, the key aspect at the initial stages of the treatment is to rapidly detect the infection with high sensitivity and specificity.134 One of the major concerns is mortality due to sepsis, whose rate is increasing, e.g. from 27% to 41% in 6 years, according to data from German hospitals.135 Neonatal sepsis, for instance, is present as a cause-specific mortality and morbidity indicator with significant contribution to the causes of death among children aged <5 years.136 One of the most common bacterial infections is caused by Escherichia coli (E. coli), which involves neonatal meningitis, bacteraemia, etc. When the condition becomes critical, the infection can evolve to haemolytic-uremic syndrome (HUS) due to E. coli in children and adults. HUS leads to haemolytic anaemia, and in severe cases to kidney failure that can cause patient death.137 The effect on public health demands continuous improvement and development of analytical devices for detection of infections. Robust, sensitive, specific and culture-free pathogen detection is necessary because there are several infections due to a single microorganism (Mycobacteria), but other practical limitations are also present. For instance, fungal pathogens may require lengthy (20 days) laboratory procedures to complete one division cycle, while there are some E. coli strains that require 20 minutes to divide. Thus, fast and reliable solutions for healthcare providers are urgently needed. This drive to decrease time of diagnosis and to improve testing accuracy could eventually reduce infection-related morbidity and mortality rates.
Conventional methods such as microbial culture-based and molecular assays (immunological – ELISA or nucleic acid technologies-based – polymerase chain reaction (PCR)) in clinical bacteremia diagnostics take between 1–2 days to reach targeted treatment. These techniques involve cumbersome sample preparation/processing (plating, growth, colony counting) and examination inside a hospital setting and still only represent an interim solution. The current limitations of these standard protocols for sample analysis and infection diagnosis are constantly challenged by trauma situations, on-site medical interventions, rapid analysis in outdoor conditions and biodefense. High-sensitivity and multistep sample processing in current diagnostic techniques demand skilled manpower, which is a barrier in the case of point-of-care (PoC) in low-income countries. However, while there are still challenges and limitations in pathogen detection that need to be resolved and overcome, notable improvement has taken place in the last decade.138 Following a general trend in healthcare, infection diagnostics are moving towards a patient-centered system that would eventually enable collection, testing and early diagnosis near the patient for a targeted treatment with no delay. Within this context, the combination of optical sensors with microfluidics is becoming ever more present and practicable. The requirements are preparation and investigation of the sample or analyte on a single device for a short analysis time, multiplexing detection for most common pathogens, automation, portability and versatility.139
Fluorescence-based sensors are still time-consuming and require external and large accessories, limiting their use in PoC clinical environments. Plasmonic biosensors are a good alternative since they provide real-time, direct pathogen detection and come nowadays in miniaturized140 or even smartphone-based portable devices.141 Biosensors with wireless communication capability are currently highly desirable for pathogen diagnostic systems because they enable remote monitoring.
Spectroscopy-based approaches are vital assets in addressing the current, main issues of pathology diagnosis. Due to limitations in infrared optics, it is quite difficult to analyze a single bacterium. By contrast, Raman microspectroscopy (RMS) is a non-destructive analytical method that uses visible light and has the ability to achieve high spatial resolution by employing high numerical aperture objectives for magnification. Conventionally, only those samples that are in bulk amount or relatively concentrated solutions have been analyzed by Raman spectroscopy due to limitations of sensitivity. However, the nanoscience phenomena involved in SERS have overcome these limitations, by allowing detection down to the level of a single molecule1,2,4 or single cell.142,143 In addition, SERS benefits from high specificity expressed through molecular fingerprinting. Additionally, by exploiting local optical fields of specialized metallic nanostructures, SERS can (in some cases) provide lateral resolutions better than 10 nm, which is a striking two orders of magnitude below the diffraction limit.144
These unique characteristics of SERS-based approaches offer promising potential in various applications including pathogen detection, high-throughput screening and infection diagnosis.145 The main advantage remains the rapid (within minutes)146,147 and reliable bacterial spectral response that is so critical in the case of sepsis awareness and prevention. For instance, diarrhoea is one of the leading causes of high mortality rates in children under 5 years of age. It is caused mainly by Vibrio cholerae and E. coli (EPEC)148 which utilize quorum sensing (an intercellular communication process) to determine their own population size. During the period of colony formation, the biofilm grows by division of existing cells and, furthermore, by attracting other microorganisms. Biofilm maturity is reached when the intra- and intercellular signalling develops. To facilitate their sensing process, bacteria synthesize and secrete signalling molecule autoinducers (AIs) in high concentration if the population size is increased. For V. cholerae, at high AI concentrations, the genes responsible for virulence and biofilm formation are repressed,149–151 as an adaptive mechanism.
However, the analysis of biofilm morphology is challenging due to their thickness; the problems are due to the limitations of surface- and bulk-specific analytical techniques. The biofilm thickness can vary widely but it is usually in the 10–100 μm range. This thickness precludes a full analysis of the surface, by surface analytical techniques. Even more challenging is the in-depth biochemical analysis of clinical samples, which demands the detection and quantification of the major pathogenic subpopulations on the biofilm. The RMS-based detection of extracellular polymeric substances (EPS) instead of bacteria reveals three main components found in biofilms: DNA, proteins and polysaccharides.152 All of these biomolecules can be fingerprinted with well-resolved bands in the SERS spectra. Moreover, the observed intensity of SERS fingerprints depends on molecular concentration, thereby providing quantitative information.
As discussed above, there are two SERS approaches that are commonly employed for bacterial detection. The first one is the label-based detection approach that applies a SERS tag as a quantitative reporter and also needs biorecognition molecules (such as antibodies and aptamers) that specifically bind to pathogens (Fig. 11). For the label-based simultaneous detection of E. coli O157:H7 and S. typhimurium, two novel SERS tags were designed and developed by Li et al.153 In the synthesis of SERS tag-1, an aptamer complementary to E. coli O157:H7 and DNTB was used to achieve a cracked octahedral shape. Similarly, an aptamer complementary to S. typhimurium and MBA was employed in the preparation of SERS tag-2 to achieve small protrusions (Fig. 11Aa). Afterwards, both aptamers along with the Raman reporter molecules were incubated with gold nanorods (GNRs). Following incubation, the process resulted in simultaneous SERS pathogen detection (Fig. 11Ab).
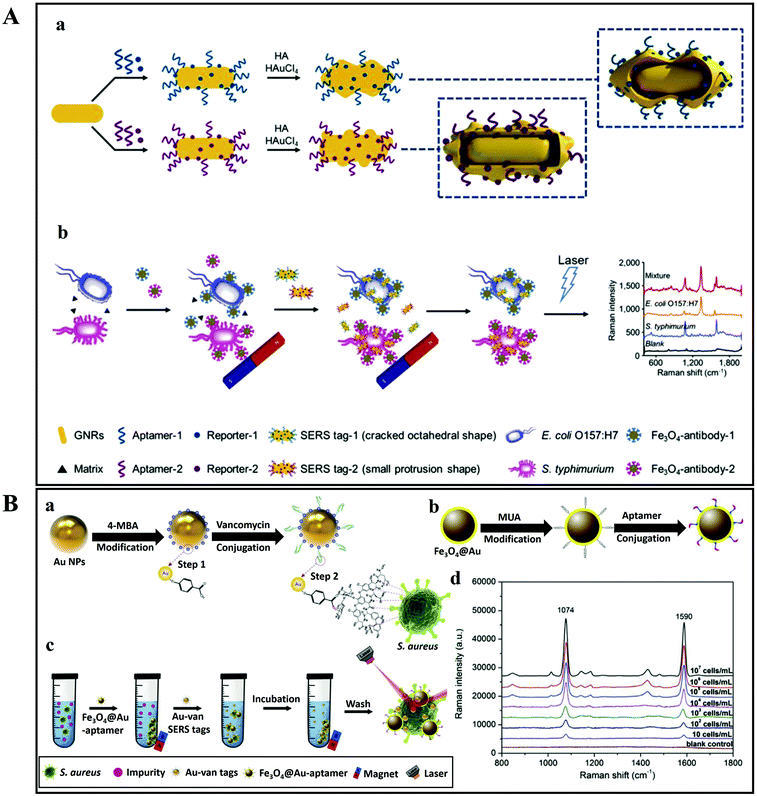 | ||
| Fig. 11 (A) Schematic illustration of the multiplex SERS detection of bacteria on gold nanorods using Raman reporters and oligonucleotide aptamers. (a) Preparation of SERS tag conjugated with aptamer and Raman reporter, and (b) separation and SERS detection of bacteria. Reproduced with permission.153 Copyright 2020, Elsevier. B.V. (B) Pathogenic bacteria detection using aptamer–Fe3O4@Au magnetic nanoparticles and vancomycin SERS tags. (a) Au–vancomycin SERS tag synthesis, (b) aptamer–Fe3O4@Au magnetic nanoparticle synthesis, (c) diagrammatic sketch of the bacterial detection process, and (d) SERS spectra with various concentrations of S. aureus (107–10 cells per mL). Reproduced with permission.154 Copyright 2019, Elsevier. B.V. | ||
In another study, aptamer–Fe3O4@Au magnetic nanoparticles (AuMNPs) were prepared as SERS substrate, and vancomycin–SERS tags (Au@MBA) were synthesized for the detection of pathogenic bacteria (Fig. 11B).154 The preparation processes of Van–SERS tags and aptamer–Fe3O4@Au MNPs is presented in Fig. 11Ba and b, respectively. Once the solution containing different species of bacteria was mixed with aptamer–Fe3O4@Au MNPs, the target bacteria were captured by aptamer recognition. This process is illustrated in Fig. 11Bc. After magnetic separation, the SERS spectra were measured. Fig. 11Bd presents the SERS spectra with various concentrations of S. aureus (107–10 cells per mL). Such SERS-based techniques can be applied for the detection of different bacterial strains with precise quantification ability by using different aptamers.
The label-based strategy is a highly sensitive approach that enables the multiplex detection of pathogenic bacteria; however, it suffers from limitations when it comes to high-throughput bacterial detection. For instance, essential biological information related to bacterial cells (physiological state, antibiotic-driven modifications of the cell wall, etc.) is lost upon using this technique. In addition, the whole procedure is laborious and time consuming as it involves secondary dye usage that results in an increased demand of reactant volume and preparatory steps, which leads to increased total analytical time.
The second SERS approach that was discussed above is label-free SERS detection that directly probes the intrinsic vibrational fingerprint of pathogenic bacteria by ensuring that the pathogen is in close proximity to the nanostructured noble metal surfaces (Fig. 12).155 In a study by Tahir et al., bioaerosol containing E. coli was detected by SERS using Klarite SERS substrate.156 The whole system involved in the label-free detection of E. coli comprises Klarite SERS substrate, bioaerosol generator, Andersen sampler and a turbulator surrounded by a closed chamber (Fig. 12A). First, a nebulizer generated bioaerosols in the chamber. Then the bioaerosols were distributed inside a chamber with a turbulator. An eight-stage Andersen sampler was placed in the chamber to collect bioaerosol on Klarite substrates. Fig. 12A shows the SEM image of Klarite with E. coli after the collection of bioaerosols. The collected airborne bacteria were analyzed with SERS. This label-free approach offers culture-free (and therefore rapid) analysis of atmospheric bioaerosol.156
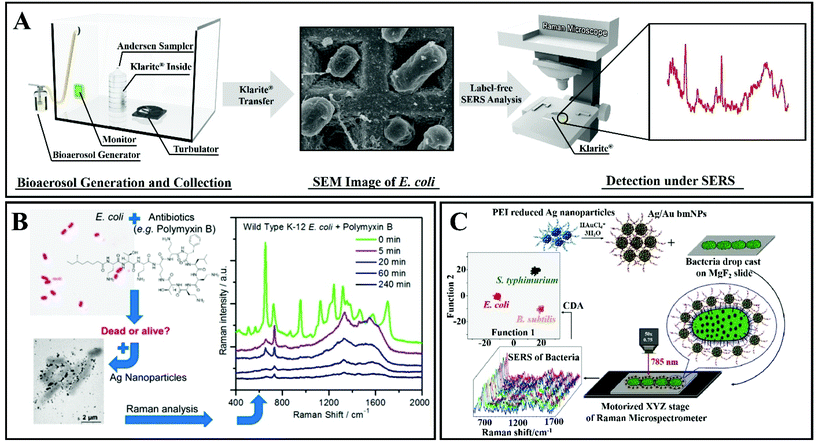 | ||
| Fig. 12 (A) Scheme of the label-free detection of E. coli by using Klarite as the SERS substrate. Reproduced with permission.156 Copyright 2020, The Royal Society of Chemistry. (B) SERS allows rapid counting and discrimination between live and dead bacteria, coated with silver nanoparticles. Reproduced with permission.157 Copyright 2015, American Chemical Society. (C) Multiplex SERS-based bacterial detection with positively charged Ag/Au bimetallic nanoparticles. Reproduced with permission.158 Copyright 2020, American Chemical Society. | ||
In another study, Zhou et al. have reported a technique for rapid counting and successful discrimination between live and dead bacteria. They used nanoparticles as SERS substrates (Fig. 12B).157 In addition, they have also confronted the bacteria (wild type K-12 E. coli) with different antimicrobial agents including polymyxin B. The SERS signal sharply decreased when the wild type K-12 E. coli was killed by 10 μg mL−1 of polymyxin B. This study not only allows us to differentiate between live and dead bacteria, but it can also differentiate between susceptible and resistant bacterial types/strains.157
Ag/Au nanoparticle-based bimetallic SERS substrates were developed for the label-free detection of E. coli, Salmonella typhimurium, and Bacillus subtilis (Fig. 12C).158 In the preparation of Ag/Au bimetallic NPs, polyethylenimine (PEI)-reduced Ag NPs were employed as a sacrificial template and AuCl4− ions were used as metal precursors. The unreacted amine groups on Ag NPs and the Ag NP colloidal solution behave as reducing agents for Au. For the synthesis of Ag/Au bimetallic NPs, the most important step is the alloying process that proceeds by surface-segregation phenomena. The positive surface charge on the Ag/Au bimetallic NPs enables the capture of the bacteria. Subsequent SERS allows accurate discrimination and classification of the three different kinds of bacteria (Escherichia coli, Salmonella typhimurium and Bacillus subtilis) with excellent reproducibility.158
The direct, label-free approach is more appealing than the indirect, reporter-mediated detection approach, as the latter is usually costly and it increases the analytical time. Consequently, substantial efforts have been directed towards developing SERS biosensors.
3.1. SERS biosensors
A biosensor is usually defined as a device combining a biological recognition element and a suitable transducer. The recognition element should interact selectively with the target analyte, and the transducer element should signal the binding event. In SERS biosensing, the unique physico-chemical properties of nanostructured silver/gold (e.g. ease of surface functionalization) are combined with the inherent enhancement in Raman signal due to the surface plasmon resonance.159 Because SERS can be applied in biofluids, it is directly applicable in clinical environments.For urinary tract infection (UTI) diagnosis, for instance, a urine culture is tested to identify germs. It takes up to 24 hours to obtain preliminary results and it takes at least 48 hours to obtain definitive results (using urinalysis, microscopy) including antimicrobial susceptibilities. During this time, an empiric suspected UTI treatment is initiated until culture results can be obtained. With the recent advancements in molecular diagnostic technology, a robust RMS-based bacterial infection diagnosis160,161 is possible, in order to complement the current culturing techniques, thereby potentially avoiding incorrect antibiotic therapy or unnecessary treatment (should there be no infection), both of which result in antibiotic resistance. E. coli, Staphylococcus aureus, Enterococcus spp., Klebsiella pneumoniae and Proteus vulgaris are the most common uropathogens in children. In the case of Enterococcus, for instance, it was demonstrated that differentiating between vancomycin-resistant and vancomycin-susceptible strains is possible162 after bacterial capturing from blood samples.163 This is a major step towards future clinical applications.
Another concern is that the number of fungal species that may develop into invasive fungal infections (IFI) is also growing with an alarming speed. Such infections can lead to deaths.164,165Candida and Aspergillus are the main invasive fungal species responsible for infections; thus candidiasis and aspergillosis are the most common fungal infections found in clinical practice, especially in immunocompromised patients (either due to a congenital immunodeficiency or due to a weakened immune system for other reasons). The classical methods of fungal infection diagnosis are blood testing, culturing (which can take from days to weeks) or using biomarkers such as galactomannan and β-D-glucan, which is a non-invasive and non-species-specific method; hence, the latter does not allow targeted treatment.
New SERS-based biosensors are being developed with a very high speed of testing in real-life applications.166–168 The advanced optical analytical techniques (e.g. assays based on fluorescent or chemically luminescent probes and gold NP-based colorimetric sensing) and particularly the SERS-based techniques for biosensing and bioanalysis are robust, fast (tens of minutes to several hours) and ultrasensitive (down to single bacteria). Currently, metallic NPs can be combined in even more efficient (higher sensitivity and specificity) nanocomposites/conglomerates with graphene oxide (GO).81 For instance, high performance SERS-active substrates were realized by directly growing graphene onto Ag NPs169 or via graphene–Ag-NP–silicon sandwich nanohybrids.170 GO is preferred for the protection against oxidation of the Ag NPs and for its lack of influence on SERS.171 A graphene monolayer covering (by chemical vapor deposition) of the Ag NPs efficiently stabilizes the SERS-active substrate, providing a month's lifetime and use under ambient conditions. Graphene-based materials in combination with Au NPs are most frequently used in biosensing for the detection of biomolecules due to their versatile functionalization chemistry.172 Using an environment-friendly method, a composite of Ag NPs and reduced GO was prepared in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. It was demonstrated that this composite allows bacterial detection.173 More specifically, it was shown that graphene is acting as a support and surfactant that can control the growth of the Ag NPs and can ultimately boost the chemical enhancement effect of SERS by providing favorable conditions for charge transfer between biomolecules and graphene sheets.174,175
1 ratio. It was demonstrated that this composite allows bacterial detection.173 More specifically, it was shown that graphene is acting as a support and surfactant that can control the growth of the Ag NPs and can ultimately boost the chemical enhancement effect of SERS by providing favorable conditions for charge transfer between biomolecules and graphene sheets.174,175
Another approach for the detection of pathogens using SERS nanobiosensors is provided by nano-sculptured thin films (nSTFs), grown via a glancing-angle deposition technique.176 These prepared chips show promising sensing power, enabling E. coli detection down to single-bacterium level. Impedimetric detection of pathogenic bacteria using peptide sensors has recently attracted interest due to its sensitivity in real-life samples.177,178 Since sensitive in situ detection of organic molecules is highly desirable, several SERS-based approaches were adopted in the microfluidics area in order to obtain high-throughput detection protocols.179
Yuan et al. found that multiple bacterial pathogens can be captured via magnetic separation and SERS tags.81 As illustrated in Fig. 13, this approach depends on the development of an Fe3O4 NPs/bacteria/SERS tags sandwich structure. First, the sample matrix with bacteria and blood cells was cultured with antimicrobial peptide (AMP)-modified Fe3O4 NPs (Fig. 13a). AMPs are widely used as capturing elements for their stability, low cost, and available recognition sites.177,182 As the AMP-based Fe3O4 NPs bonded with the bacteria, it was possible to magnetically separate the bacteria (Fig. 13b) from the rest of the sample matrix (Fig. 13c). A sandwich-type structure formed after incubating the 4-mercaptophenylboronic acid (4-MPBA) and Raman reporter molecules (Fig. 13d). A boronic acid group is present in 4-MPBA, which helps in binding to the cell wall of bacteria using peptidoglycan. Finally, magnetic separation allows the sandwich structure to be collect for SERS analysis (Fig. 13e). 4-MPBA is a reporter molecule in this study and it can discriminate between different kinds of bacteria by modifying their Raman “fingerprints” (Fig. 13f). In addition, SERS intensities can be normalized by using 4-MPBA as an internal standard (Fig. 13g).81 Therefore, this SERS sandwich-based approach can be applied to the sensitive and selective determination of pathogenic bacteria.
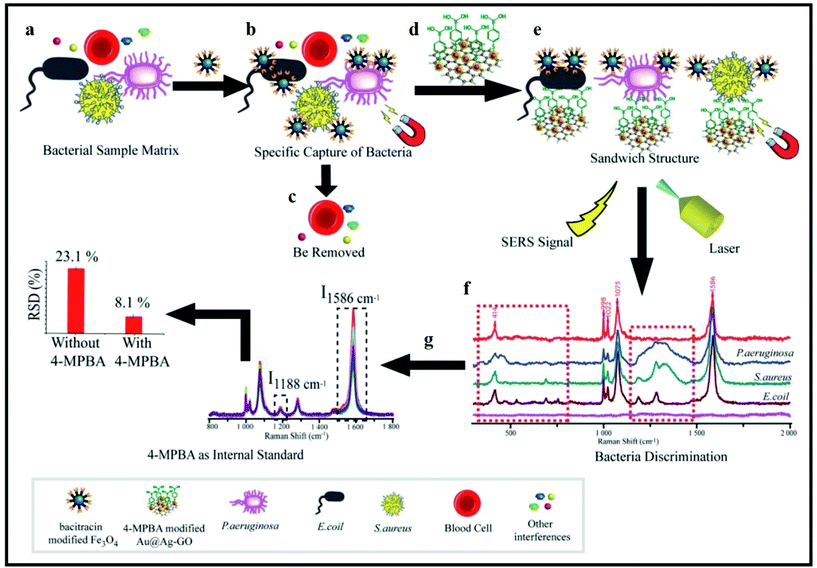 | ||
| Fig. 13 Schematic illustration of bacterial detection using a SERS sandwich strategy based on magnetic nanoparticles functionalized with antimicrobial peptides for the separation of bacteria, and a SERS tag of 4-mercaptophenylboronic acid (4-MPBA)-modified gold-coated silver-decorated graphene oxide (Au@Ag–GO) nanocomposites. (a) Sample matrix was cultured with antimicrobial-peptide-modified Fe3O4 NPs. (b) Magnetic separation of the Fe3O4 NPs@bacterial complex from the sample matrix. (c) Removal of blood cells including other interfaces. (d) Sandwich-type structure formed after culturing the Fe3O4 NPs@bacteria with Au@Ag–GO nanocomposites. (e) Magnetic separation of the Fe3O4 NPs/bacteria/SERS tag sandwich structure. (f) Discrimination of different kinds of bacteria with SERS. (g) SERS intensities can be corrected by using 4-MPBA as an internal standard. Reproduced with permission.81 Copyright 2018, The Royal Society of Chemistry. | ||
Recent progress has been reported in using SERS in subcellular compartment research, for elucidating pathological mechanisms that might improve the early diagnosis and therapy of modern diseases.183In situ visualization of cellular organelles by targeting nanoprobes offers 3D imaging with high resolution of single cells; mostly the cell membrane and the nucleus are being investigated. Nanomaterials enter cells through the clathrin-mediated endocytosis pathways, accumulate in lysosomes or cytoplasm and are used for imaging or photothermal therapy. Also, the benefits of dual-recognition SERS-based approaches for pathogenic bacteria detection are increasingly recognized; these include superior specificity and sensitivity, minimized matrix interference and reduced costs when using non-specific recognition elements (antibiotics, bi-functional small molecules, etc.).184
3.2. Spectral data intelligent analysis by using unsupervised and supervised multivariate analysis methods
Spectroscopic techniques require instrument calibration using a chemical reference. Moreover, decoding the spectral information presents difficulties in data handling, as overlapping spectra and complex mathematical operations need interpretation. Consequently, the operation of such equipment demands the employment of highly skilled personnel.However, following technological progress, spectral acquisition times have been reduced to (a maximum of) several minutes for hundreds of spectra. For bacterial spectral fingerprinting and classification, it is still challenging to perform rapid and intelligent data analyses. Innovative detection and molecular diagnosis tools are now integrated with Raman spectroscopy and nanotechnology,162,185–188 assisted by effective chemometrics.
Chemometrics has already been integrated with spectroscopic laboratory and process instrumentation and has proved effective in facilitating spectral data analysis by employing principal component analysis (PCA), partial least squares (PLS) regression, linear discriminant analysis (LDA) or cluster analysis.189–191 Following a reduction in the post-processing steps of data analysis, a dramatic increase in label-free SERS detection usage can be expected. At the moment, the extraction of the large volume of spectral information still requires complex multivariate analysis. The common analysis approach of the Raman/SERS spectral profiles is to look into the changes in intensity of individual bands. However, this approach can only provide semi-quantitative information.
Raman spectra of biological samples often exhibit variations originating from different-generation spectrometers, measurement parameters, and cultivation conditions. Such undesired variations can distort the classification model, especially if they are more significant compared with the differences between the groups to be separated. A classifier is prone to these intragroup variations and can fail to learn the patterns that help separate different groups (i.e. intergroup differences). Support-vector machines192 and complex data processing193,194 are able to provide remarkable results by using dedicated software. On the other hand, the multi-level computational models (PCA–PLS–LDA or PLS–LDA–LDA) that are checked and validated by using data from an independent biological replicate produce high sensitivity and even higher specificity with respect to the prediction based on minor spectral changes.162,192,195
Recently, unique rapid chemometric techniques and models were employed based on fuzzy set theory to achieve robust and high-precision fingerprinting and classification of pathogens.48 Fuzzy methods present several advantages over standard methods. For instance, fuzzy clustering methods can identify outliers (as exception data, as new trends, or somehow interesting data items), hybrid data and noise data. Moreover, they affirm the fuzzy prototyping as meaningful data items, or the divisive cluster substructure, serving as a final fuzzy hierarchy and as inner substructure data, as class relationships. Fuzzy hierarchical cross-clustering (FHCC) algorithms provide a better association between each set of samples to a fuzzy set of characteristics. Moreover, FHCC techniques are unique due to simultaneous clustering of objects (samples) and characteristics (variables) but also the 3D biplot option according to the membership degrees of samples and variables to the final fuzzy partitions. Additionally, for fuzzy PCA/LDA methods, a better compression, robustness towards outliers and more accurate data separation were demonstrated.196–200 The main issues solved by employing fuzzy algorithms (fuzzy divisive hierarchical clustering, fuzzy cross-clustering, fuzzy PCA,201,202 fuzzy LDA203) are (i) classification and fingerprinting of spectral data40,204–206 and (ii) discrimination of multi-resistant common strains found under antibiotic/antifungal stress/susceptibility conditions.48 Due to the original and complex approach, the results on SERS data from bacteria are scarce. The fresh impetus towards this research direction is the recent development in SERS chemical imaging of live cells. Practically, a SERS image can be generated within few minutes followed by a rapid multivariate cluster analysis: a very useful approach for real-time physico-chemical processes monitoring at the plasmonic-membrane interface or at intracellular level.207
4. Point-of-care (PoC) solutions based on SERS for infection diagnosis and targeted treatment
The emerging high-throughput diagnostic methods allow a quick testing of DNA208 and antibody arrays.209 The reduced cost and time of an assay and the long storage feasibility make SERS a strong candidate for large-scale sample screening in clinical applications.210 SERS can serve to rapidly sequence DNA without the need for fluorescent labels.207,211–215Antibiotic multiresistance (AMR) is still tested in hours-to-days time periods, by using plating and costly susceptibility tests. Molecular biology specific techniques are capable of finding the genes responsible for the multiresistance and generate valuable input in the case of generalized infections, but with important time costs. Implementing SERS as a tool for spectral pathology or even for rapid targeted treatment identification is yielding promising results in the clinical environment. In order to control antibiotic misuse and handle the multi-drug resistance issue, it is important to investigate bacterial antibiotic susceptibility. Ideally, real-time pathogen susceptibility measurements without labelling or other complicating pretreatment of the sample would be performed. Recent studies in combined dielectrophoresis (DEP)–Raman spectroscopy seek to identify the simplest, most reliable and fastest approach.216 Schröder et al.162,217 and Kirchhoff et al.218 also proposed spectroscopy-based methods for antibiotic susceptibility testing (AST). For infection diagnosis and sepsis prevention, SERS-based characterization and rapid identification of the main pathogen would be highly beneficial.48,52,219
Recently, a rapid and sensitive SERS-based AST technique was established by using bacteria–aptamer@Ag NPs.220 The minimum inhibitory concentration (MIC) was determined in approximately 1 hour and the representative tested pathogens showed a similar spectral profile in the case of sub-MIC antibiotic concentrations: a significant increase of the 735 cm−1 SERS band after 1 hour. Thus, SERS can be applied as a highly sensitive in situ fingerprinting tool to assess antibiotic susceptibility both on Gram-positive and Gram-negative pathogens by employing carefully screened and selected aptamers. This SERS–AST approach proved to be a rapid and reliable way of detecting live bacteria by assessing the evolution of the most intense SERS band (735 cm−1) as a function of the antibiotic concentration applied and for monitoring how sub-MIC antibiotic pressure is leading, as demonstrated, to increasing bacteria colony-forming units (CFUs).220
Another noteworthy and recent Raman-based approach for robust and valid antibiotic susceptibility testing of bacterial pathogens was reported by Bauer et al.221 Adding stable isotope labelling in Raman-based antibiotic susceptibility testing further improves the sensitivity. A protocol for both Gram-positive and Gram-negative pathogens was assessed for Raman-based AST within 3 and half hours, including sample preparation, measurements, and analyses. Bacterial interaction with the NPs starts at the NPs’ surface. For instance, silver-based NPs discharge silver ions that attach to biological binding sites upon surface adhesion, performing an antibacterial activity. However, there is very little knowledge regarding the correlation between the surface properties of bacteria and the nanosilver/nanomaterial's antibacterial mechanisms. Recently, it was reported that biofilm formation is resisted by silver-doped laser-induced graphene-coated surfaces,222 with potential use in the decontamination of rural water sources or in the case of disaster situations. So far, the capacity to investigate the interfacial interaction phenomena in detail by employing in situ techniques appears limited.223 However, in situ SERS analysis and barcoding can retrieve key information about the origin of the observed SERS bands, their accurate assignments and a better understanding of the interaction between silver nanoparticles with the bacterial cell wall.224
Despite the fact that SERS can be regarded as a highly sensitive detection technique, especially in the case of biological samples, the reproducibility of SERS spectra, when performing single-cell SERS measurements, is still low. There are two main ways to prepare samples for SERS bacterial detection using colloids as active substrates: either the nanoparticles coat the outer cell wall of the bacteria or they are directed inside the bacterial cell. In the former case, the SERS spectral information originates from cell wall components, while in the latter case the cytoplasm is being probed. Thus, SERS spectra can comprise both intra- and extra-cellular information, which improves specificity. The bacterial outer membrane's chemical and biological components carry molecular specificity pertaining to the strain, growth phase and metabolic state. This biochemical information was used from seven strains of V. parahaemolyticus for designing a SERS barcoding model system able to detect and identify “blind” real samples even in mixtures.225
SERS spectra are not straightforward to interpret. Hence, spectral fluctuations caused by the drying process should be avoided by using, for instance, miniaturized sealed chambers (microchannels, capillaries, etc.) as a detection window or by recording spectra through a protective water layer. In addition, spectral information derived from bacterial biomass should be collected as accurately as possible (without contributions from the matrix of extracellular polymeric substance (EPS) species) by implementing purification or selective preconcentration steps. SERS signals are usually linked to cell wall contributions such as modes of flavin derivates and of polysaccharides.226,227
Some strategies for overcoming the technical drawbacks of single-bacterium SERS detection are pointed out in the following section. A common challenge of SERS-based assays for detecting and analyzing bacteria is to design and prepare such techniques that are capable of capturing bacteria from water, phlegm, saliva or blood samples and to directly load them onto SERS substrates without compromising their SERS-sensing capability.228
4.1. SERS-based methods for bacterial infection and disease diagnosis
In this sub-section, the main two approaches of label-based and label-free detection of biomolecules in spectral pathology are surveyed, within the context of their associated clinical perspectives.Xu et al. have reported a SERS substrate comprising Au NPs functionalized with single-stranded DNA (aptamers) that acts as a capture probe.230Fig. 14A illustrates the preparation of asymmetric gold nanodimers (Au NDs) and working principles for the detection of S. typhimurium. The Au NPs bind with thiol-modified single-stranded DNA-1 (ssDNA) to produce Au–S bonds. The signaling probe comprises Au NPs conjugated with the aptamer and with its complementary Cy3-labeled strand. Subsequently, in the presence of S. typhimurium, the aptamer releases the Cy3-labeled strand and binds to the S. typhimurium. It is then possible to use a Millipore filter, in order to separate the S. typhimurium (with its capture probe). Finally, the filtrate is studied with SERS to analyze the S. typhimurium concentration.230
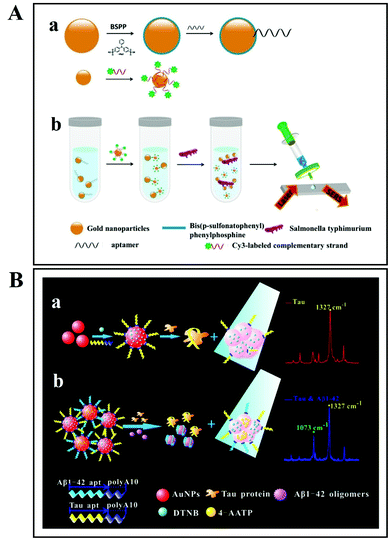 | ||
| Fig. 14 (A) Schematic representation of gold nanodimer (AuNDs)-based aptamer for bacterial detection. Reproduced with permission.230 Copyright 2018, Springer Nature. (B) Schematic of the detection process of Alzheimer's disease biomarkers: Tau protein and Aβ(1–42) oligomers using Tau–aptamer–polyA10–Au NP and Aβ(1–42)–aptamer–polyA10–Au NP conjugates respectively. Reproduced with permission.231 Copyright 2019, American Chemical Society. | ||
Similarly, a SERS sensing platform based on dye-coded polyA aptamer–gold nanoparticles (PAapt–Au NPs), Aβ(1–42) oligomers and Tau protein for sensing Alzheimer's disease (AD) biomarkers is reported by Zhang et al.231 (Fig. 14B). The PAapt–Au NPs conjugates are formed after the self-assembly of polyA block nucleotides and nonfluorescent Raman dyes on the Au NP surface. The polyA block helps in attachment to the surface of Au NPs, while protein recognition is performed by polyA bock nucleotides comprising oligonucleotides. After addition of a protein biomarker in the reaction system, the polyA block nucleotide detaches from the Au NP surface, following hybridization of the target protein with its specific aptamer. Then, aggregates of Au NPs are formed resulting in enhanced electromagnetic hotspots (within the gaps between nanoparticles) and in a significantly stronger SERS signal. However, there is also a relatively weaker SERS signal in the absence of protein biomarker (Fig. 14B). This approach depends upon the protein–aptamer binding-mediated aggregation of Au NPs and the plasmonic coupling effect that will eventually help in detecting the protein biomarkers via SERS.231
As a single-cell approach to investigating the metabolic activity of bacteria, recently an elegant way of using Raman stable isotope labelling with 18O has been reported.232 Raman 18O labelling is used by exciting with a non-resonant 532 nm wavelength to monitor the incorporation of 18O in proteins and with the resonant 244 nm UV excitation for information specific to the nucleic acids of the bacterial cells. The spectral differences between labelled and non-labelled E. coli cells for the same excitation wavelength were only highlighted by the PCA–LDA analysis. Specific for isotope metabolic assimilation, amide band I peak shifted to lower wavenumber positions when exciting at 532 nm and its intensity decreased in time. A 2D correlation analysis was applied for representing synchronous and asynchronous 2D spectra in the range 1750–1520 cm−1 as time-dependent spectral variations for E. coli cells. The patterns obtained were only specific to labelled cells, suggesting variations due to the incorporation of the oxygen isotope. By this approach, access to physiological and genetical information about bacterial cells is obtained.
An interesting “Click” SERS readout platform was also reported.233 This approach comes as a solution for the SERS detection that is label-based and with a single readout mode, still suffering from difficult quantitative sensing. This novel strategy is based on the controllable splicing of layers from modified Au NPs containing DNA and sequential DNA hybridization in the presence of caspase-3 as a trigger. In detail, polyA–DNA and Asp–Glu–Val–Asp (DEVD)-containing peptide sequence were used for modified Au NP preparation. The prepared Au NPs with complementary DNA sequences and Raman reporters were incubated with caspase-3 and used after reaction completion (Fig. 15a–c).
 | ||
| Fig. 15 Scheme of the “Click” SERS detection for caspase-3. (a–c) Reaction principle between peptides and caspase-3: polyA–DNA and DEVD-containing peptide sequence were used for modified Au NP preparation. The prepared Au NPs with complementary DNA sequences and Raman reporters were incubated with caspase-3. Caspase-3 leads to cleavage of the DEVD peptide, removing the protective layer from the surface of the Au NPs. The labelled Au NPs are connected via DNA hybridization and form SERS-active “hot spots” for detecting the Raman tags. The Raman lines found at 2158 cm−1 for OPE1 and at 2227 cm−1 for MBN are the “Click” SERS signals used as readout. Reproduced with permission.233 Copyright 2021, American Chemical Society. | ||
During cellular apoptosis, caspase-3 release leads to the cleavage of the DEVD peptide, removing the protective layer from the surface of the Au NPs. The labelled Au NPs are connected via DNA hybridization and form SERS-active “hot spots” for detecting the Raman tags. Alkyne- and nitrile-terminated molecules (MBN and OPE1) were chosen as artificial Raman signals for dual simultaneous detection – the “Click” SERS readout. They present strong and sharp signals in the cellular-silent region (1800–2800 cm−1). The Raman lines found at 2158 cm−1 for OPE1 and at 2227 cm−1 for MBN are the “Click” SERS signals that allow single-cell SERS imaging. Following an apoptosis-related mechanism induced with doxorubicin, SERS measurements of caspase-3 activity in vitro (in HeLa cells) were performed. For this, the linear relationship between the SERS marker band intensities of the two selected Raman tags and the concentration of caspase-3 was proved. The “Click” SERS readout was significantly reduced when the cells were incubated with a caspase-3 inhibitor, supporting the probe's high specificity in imaging. The “Click” SERS strategy shows zero noise background for single-cell SERS imaging and could be applied for enzymatic activity assays and living cell bioimaging.
A clinical-sample-oriented biochip for bacterial capturing and their in situ detection in human blood was developed by Liu et al. (Fig. 16A).228 By growing Ag NPs on arrays of anodic aluminum oxide (AAO) nanochannels to take advantage of the sub-10 nm inter-particle gaps, they created ‘hot junctions’ for the electromagnetic enhancement of Raman signals. During SERS analysis, bacteria from blood samples were concentrated on a vancomycin-coated surface, as there is specific binding between vancomycin (Van) and the bacteria. Van-functionalization also helps in reducing the adhesion of the blood cells and biomolecules in the blood onto the SERS substrate (Fig. 16A). Bacterial capturing was achieved both from blood and from water, by hydrogen bonding of the peptidoglycan species within the bacterial cell wall with the amine and carbonyl groups of the Van-coated Ag/AAO arrays. Due to the bacterium-binding capacity of the Van-coated substrate, it is important to check whether the Van susceptibility test can also be carried out for different strains of Gram-positive bacteria. Importantly, upon using the designed Van-coated substrate, a distinct SERS signal can be observed from Van-susceptible Enterococcus (VSE), as compared with Van-resistant Enterococcus (VRE). Thus, a SERS-based in situ detection method with a high performance in sensing and monitoring the response to antibiotic treatment was obtained.228 These SERS results exhibit the feasibility of the proposed Van-coated substrate for antibiotic susceptibility testing (AST). Furthermore, SERS along with control nanogap chemistry and machine learning algorithms were applied for performing spectral data analysis in order to correlate the bacterial metabolic profile post antibiotic exposure with the susceptibility. With the help of deep neural models, the response of two different bacteria, Escherichia coli and Pseudomonas aeruginosa, to antibiotics from untreated cells in SERS data can be differentiated and discriminated in 10 min with 99% accuracy after antibiotic exposure.235 The high accuracy and sensitivity makes a SERS + deep learning approach an effective technique for the rapid AST in clinical samples.
 | ||
| Fig. 16 (A) Label-free bacterial detection on vancomycin-coated Ag/AAO SERS substrate. (a) Schematic representation of bacterial detection on SERS substrate, (b) SEM micrograph of SERS substrate with bacteria, and (c) optical images and SERS spectra of bacteria from human blood samples. Reproduced with permission.228 Copyright 2011, Macmillan Publishers Limited. (B) Schematic illustration of lung cancer biomarker detection via gold superparticles coated with ZIF-8 shell. (a) Assembling of Au NPs into gold superparticles (GSPs). (b) Surface of GSPs coated with ZIF-8 shell. (c) Detection of volatile organic compounds (VOCs) by SERS. Reproduced with permission.236 Copyright 2017, John Wiley & Sons. | ||
Moreover, Qiao et al. have developed a SERS substrate made from gold super particles (GSPs) coated with a ZIF-8 shell for diagnosing early-stage lung cancer (Fig. 16B).236 Gaseous aldehydes act as a lung cancer indicator that are released due to tumor-specific tissue composition and metabolism. Through a ZIF-8 channel, these gaseous aldehydes land onto the surface of SERS-active GSP substrate. The substrate is then analysed with SERS. This substrate's main application is to detect volatile organic compounds that serve as biomarkers for early lung cancer detection (Fig. 16B).236 Most recently, blood cells and circulating tumor cells (CTCs) could be rapidly discriminated by employing label-free SERS and deep learning algorithms.237 The SERS spectra obtained from eight kinds of tumor cell line were classified by using the characteristic peak ratio method, and the PCA-K-nearest neighbour (KNN) algorithm was applied for tumor cell and blood cell discrimination. Furthermore, the SERS spectra were used for training and testing a ResNet model constructed by the authors. With an identification accuracy of 100%, this label-free detection method for CTCs is an important reference study in the SERS-based diagnosis framework.
The direct approach also enables multiplexing detection assays, common in monitoring biological processes. For instance, a novel SERS probe based on hybrid graphene oxide and popcorn-shaped Au NPs was proposed for sensing HIV DNA and bacteria by Ray's group.238 Graphene oxide (GO) is preferred for its surface functionalizing ability and high potential of chemical enhancement (CM) that arises from the Raman active molecule–graphene interactions. An aptamer APTSEB1 modified hybrid GO was successfully used for selective detection of bacteria, particularly methicillin-resistant Staphylococcus aureus (MRSA). By combining the hybrid SERS probe with good affinity and high surface area (a gold nano-popcorn-shaped design), sensitive and selective direct DNA and bacterial detection is provided. The high plasmon enhancement attached to graphene has a demonstrated CM effect due to the π-conjugation network. The detection system has a long shelf life due to the highly chemically stable GO.
As an alternative, Graham et al.21 used small molecules (spermine) with positively charged functional groups as an aggregating agent in DNA detection with metal nanoparticles. The positive charge on these small molecules served to overcome the electrostatic repulsion that is caused by the negatively charged DNA phosphate groups and the metal surface. The overall particle stability in the system was improved by the presence of a ligand shell, which is particularly favourable for DNA detection.21
SERS-based protein analysis is a promising approach because it has the ability to reveal the structural changes and concentration of diverse target species; these are useful in early-stage disease diagnosis and clinic therapy.239 Park et al. have reported a carboxylic-acid-functionalized, graphitic nanolayer-coated three-dimensional SERS substrate (CGSS) made of gold nanowires for diagnosing the Alzheimer disease biomarkers tau protein and amyloid β (Fig. 17).240 The analytes used in this study were tau protein and amyloid β, which are linked with Alzheimer disease, which causes dementia. As these proteins are present in the body fluids, they are good analytes for Alzheimer disease biomarkers (Fig. 17A). In the study, a non-functionalized 3D SERS substrate was prepared by solvent-assisted nanotransfer-printing, whereby each new layer is perpendicularly stacked on the underlying layer. Fig. 17B shows scanning electron microscopy images of the 3D SERS substrate that consists of stacked Au nanowire array sheets. The SERS spectra of tau protein and amyloid β (in Fig. 17C and D, respectively) demonstrated promising prospects in secondary structure analysis and concentration measurements.
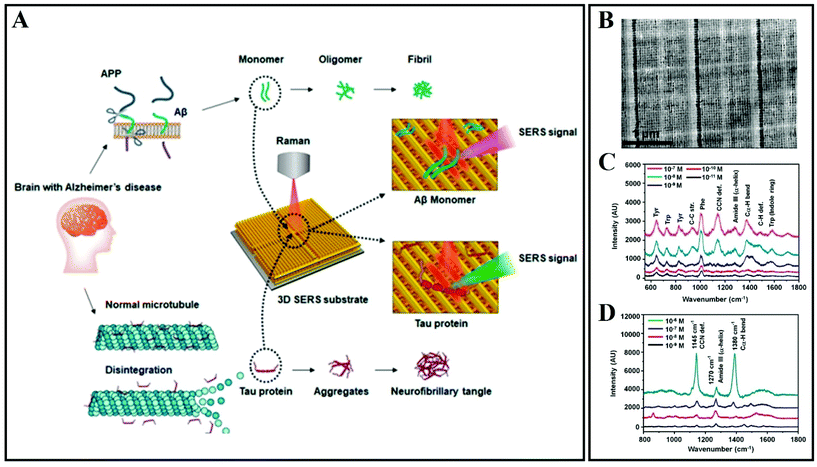 | ||
| Fig. 17 (A) Schematic illustration of label-free direct detection of Alzheimer's disease biomarkers via a three-dimensional SERS substrate coated with a graphitic nanolayer and carboxylic-acid-functionalized layer. (B) SEM image of a three-dimensional SERS substrate consisting of two or four Au nanowire array sheets. SERS spectra with various concentrations of Alzheimer disease biomarkers: (C) tau protein and (D) amyloid β. Reproduced with permission.240 Copyright 2020, American Chemical Society. | ||
Metabolomics provides information on the functionality of cells. Single-cell metabolomics reveals how cells deal with chemical or environmental stress. However, there are several limitations (including structural diversity, inability to attain labelling or signal amplification and robust metabolic dynamics) that complicate the measurements and analysis of small-molecule metabolites. To cope with this problem, Sun et al. have developed a SERS-microfluidic droplet platform as shown in Fig. 18. The label-free simultaneous detection of three metabolites at the single-cell level was achieved using a versatile magnetic SERS-active substrate, composed of Ag NPs decorated with magnetic Fe3O4 microspheres by applying PEI as a linker.241 Using separate microfluidic channels, a single cell is combined with the Fe3O4 on Ag NPs (Fe3O4@AgNPs) inside a water-in-oil droplet. Cellular endocytosis (even during long-term off-chip cell incubation) can be avoided because Fe3O4@AgNPs have a rather large size. There are different metabolites secreted by the cell during off-chip cell incubation. Such metabolites are directly adsorbed on the surface of Fe3O4@AgNPs, and then collected in droplets because of the magnetic aggregation caused by Fe3O4. Thus, SERS data are only recorded from the metabolites adsorbed on Fe3O4@AgNPs (once the metabolites’ dynamic adsorption rate attains an equilibrium). With the help of this technique, three metabolites, namely ATP, lactate and pyruvate, are detected from single cells.241 This SERS-microfluidic-based droplet platform is a useful technique to investigate and analyse the heterogeneity of metabolism of a single cell. Due to its effectiveness in single-cell metabolomics studies, this platform can be applied to study different cellular models.
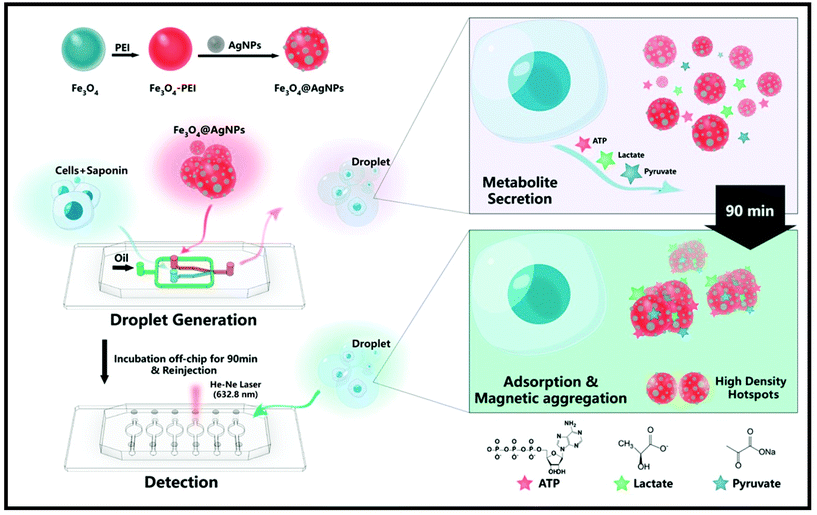 | ||
| Fig. 18 Schematic diagram of the SERS-microfluidic droplet platform for the encapsulation of single cells and label-free simultaneous detection of three metabolites (ATP, lactate and pyruvate) at the single-cell level by utilizing versatile magnetic SERS-active substrate (Fe3O4@AgNPs). This approach allows the efficient adsorption of different metabolites secreted by the cell during off-chip cell incubation on the surface of Fe3O4@AgNPs, and high SERS sensitivity. Reproduced with permission.241 Copyright 2019, American Chemical Society. | ||
| SERS-based direct approaches | Performance | Origin of SERS marker bands | Ref. |
|---|---|---|---|
| In situ detection protocols | Detection limits/EF (pros vs. *cons) | ||
| Positively charged Ag NPs+ self-assembled on the surface of the fungal cell wall via electrostatic aggregation (CTAB used as cationic surfactant) | Better than the standard substrate negatively charged Ag NPs (higher EF) | Cell wall components | 242 |
| In situ coating of bacteria/fungi with Ag NPs in two steps synthesis | Works for both eukaryotic and prokaryotic cells; discrimination between live and dead bacteria | Cell wall components | 48, 49, 52 and 157 |
| Ag dendrites mixed with bacteria (both detection and quantification via mapping technique) | *Slower than SERS single-cell approach due to mapping times required | Cell wall components | 143 |
| Capped and uncapped generated Ag NPs for bacteria interaction and detection | By using one or another synthesis method the inner or outer cell membranes are explored | Inner or outer cell membrane components | 251 |
| “Flower” shaped Ag NPs incubated with bacteria, fungi and pollens as bio-aerosol threats | *Slower than SERS single-cell approaches due to incubation times required | Cell wall components | 252 |
| Resonance Raman of cytochrome C found in A. hydrophilia | Useful for the prevention of bacterial degradation in animal skins | Metabolized components | 253 |
| A combined dielectrophoresis (DEP) – Raman spectroscopy (RS) in-house built device | AST real-time measurements | Whole cell components | 216 |
| Preincubation with antibiotic and further with D2O | Raman-based protocol AST | Carbon–deuterium stretching vibration monitoring | 221 |
| *90 minutes preincubation time prior to analysis | |||
| Ag NPs synthesized by green microwave heating technology | Quantitative detection of viable cells from dead cells | Cell wall components | 254 |
| ISO-SERS method for bacteria identification using Ag@FTO SERS substrates | 98% of accuracy in identification of food-borne pathogens in only 48 h | Cell wall components | 255 |
| *Cultivation on agar with selective media was needed | |||
| Solid plasmonic substrates | Detection limits/EF (pros vs. *cons) | ||
| *High costs and difficult scale-up | |||
| Silicon wafers covered with Ag by etching and physical vapor deposition (PVD) sputtering | High reproducibility in spectral fingerprinting; single-cell limit of detection | Cell wall components | 210 |
| Silicon wafers covered with Al either by thermal evaporation or by sputter coating and Raman microspectroscopy | Identification of viable mycobacteria in biofluids by paraformaldehyde (PFA) fixation; single-cell detection | Whole cell components | 134 |
| A combined dielectrophoresis (DEP) – Raman setup | AST and MIC testing within 2 h confirmed also by commercial Liofilchem® MIC test strips | DNA bands affected by ciprofloxacin | 218 |
| Bioinspired Ag brochosomes deposited on Au film | R6G molecules limit of detected 1 nM; bacteria concentration detected 103 CFU mL−1 | Cell wall components | 256 |
| SERS-based indirect approaches | Performance | Origin of SERS marker bands | Ref. |
|---|---|---|---|
| Specific receptors mediated protocols | Detection limits/EF (pros vs. *cons) | ||
| Antibody/aptamer functionalized Ag NPs | Real human fluids infected with very low bacterial concentration (<15 CFUs mL−1) were tested and with short acquisition time (10 min mL−1). | 4-Mercaptobenzoic acid (MBA) | 257 |
| *High costs | |||
| Aptasensor–silicon nanohybrids SERS chip (G@Ag NPs@Si) fabricated by etching chemical method and protected by graphene | Rox dye-labeled ATP aptamer | ATP | 170 |
| *High costs | |||
| Bacteria–aptamer@Ag NPs | Huge enhancement factor; MIC and AST testing in less than 2 h | Cell wall components | 220 |
| *High costs | |||
| Au@Ag core–shell nanorod (Au@AgNR) tag systems based on streptavidin–biotin functionalization | 104 CFU mL−1 (diagnostic threshold); AST potential tested; high reproducibility, specificity and bacterial capture efficiency | SERS nanotags which can specifically anchor onto E. coli cell wall | 258 |
| Lectin functionalized magnetic NPs for bacterial capture and isolation | Single pathogen detection; multiplexing nano-biosensor capability | SERS-active NPs functionalized with antibodies | 259 |
| Antimicrobial peptide functionalized magnetic NPs and Au@Ag–GO based SERS tags | 97.3% accuracy in real blood samples; multifunctional biosensing (isolation, discrimination and killing) | SERS spectra of 4-MPBA–Au@Ag–GO | 81 |
| *High costs | |||
| Novel bacitracin-based flocculant 4-MPBA modified GO–Au@Ag NPs | 103 CFU mL−1 limit of detection | SERS spectra of four bacteria with labels, SERS spectra of labels, SERS of bacteria without labels | 260 |
| Efficient sterilization mechanism | |||
The current SERS-based detection approaches are focused on recording spectra from the pathogen's membrane components, either in situ or with NPs positioned in close proximity to the cell wall.48,52,219,223,242 SERS has also been used to sense DNA,214 RNA,150 autoinducers,148 pyocyanin-like byproducts243 or mitochondrial activity indicators. Much less attention has been devoted to the extracellular products released by bacteria, in the matrix surrounding the biofilms. Yet these extracellular products can be of significant interest. For instance, saccharides could either be exploited as alternative bioresources or used to better comprehend the survival and proliferation mechanisms found in biofilms. Coupling of thin-layer chromatography (TLC) and SERS should also be propitious for on-site detection considering the simple, rapid, high-sensitivity approach.244 TLC has significant advantages over other commonly used techniques like gas chromatography–mass spectrometry (GC–MS)245,246 and liquid chromatography–mass spectrometry (LC–MS).247,248 Since first reported by Hezel,249 the TLC–SERS approach has been applied in the detection of various analytes, including biomarkers in biological samples.99,250
Advancements in nanoscience and biotechnology have enabled accurate tumor detection. Lv et al. have devised a SERS-based technique to detect cancer cells in vivo (Fig. 19).261 An environmentally friendly synthesis approach was adopted to obtain Au@Cu2−xS core–shell NPs that were later modified with folic acid (FA). There was an increase in SERS effect observed due to the integration of the plasmonic Au core and Cu2−xS shell into a single unit. A Raman reporter molecule (cresol violet acetate, CVa) was bound on the surface of the Au metal core, where it emitted a strong SERS signal. For the successful discrimination between cancerous cells and normal epithelial cells, both in vitro and in vivo, a common tumor cell membrane-specific FA receptor was conjugated on the surface of the Au@CuS core–shell NPs: Au@CuS–FA core–shell NPs. These SERS-guided NPs identified cancer cells based on their specific, strong and stable (3S) signals. Thus, FA receptor-based Au@CuS core–shell NPs not only enhanced the tumor imaging and identification, but were also involved in nondestructive photothermal therapy by 808 nm laser irradiation in vivo.261
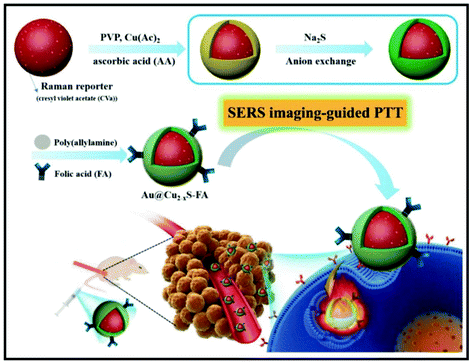 | ||
| Fig. 19 Schematic of the development of Au@Cu2−xS–FA core–shell NPs and their in vitro and in vivo SERS application for identification and photothermal therapy of cancer cells at the cellular level. Reproduced with permission.261 Copyright 2018 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
In real-time clinical applications, endoscopic imaging is a technique that allows examination of the interior cavity and hollow organs of the body, with reduced invasiveness compared with traditional surgical approaches. It is a suitable technique to screen cancer in the gastrointestinal (GI) tract. Detecting anomalies on a healthy luminal surface (the lining of the GI tract), greatly depends upon the expertise of the technician that operates the GI endoscopy instrument. It is challenging to reliably detect precancerous lesions in the stomach and esophagus. In order to address this problem, Garai et al. have reported a compact Raman imaging device based on fiber-optics to comprehensively image the lumen of a hollow organ (Fig. 20).262 By using this approach, it is also possible to analyze the SERS NPs in clinical models. During the endoscopic retraction, circumferential scanning can be applied to map the signals from SERS NPs located on the surface of the lumen (Fig. 20a). A brushless DC motor was attached to the rotating mirror that allows the circumferential scanning of the tissues over 360 degrees. In addition, a 785 nm continuous wave (CW) laser light was directed along the single-mode fiber situated in the center of the fiber bundle (Fig. 20b). Following illumination of the lumen, a scan mirror redirected the collected signal along the optical return path. There were 36 separate multimode fibers that guided the Raman scatter light to the spectrophotometer. The obtained Raman spectra were unmixed by utilizing a hybrid algorithm in combination with PCA and least squares analysis. This approach improves the diagnosis and could identify small lesions that are difficult to discern with white-light endoscopy.
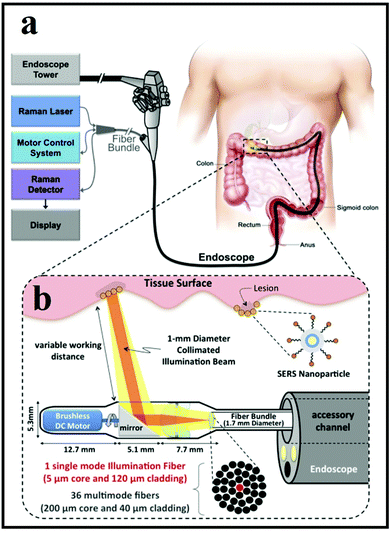 | ||
| Fig. 20 Schematic representation of a Raman-imaging system in combination with white-light endoscopy. (a) The design and construction of the device allows it to pass through the accessory channel of a clinical endoscope. (b) An expanded view of the distal end of the device. Reproduced with permission.262 Copyright 2015, Garai et al. | ||
The endoscopic SERS approach can be further improved by developing a SERS tag that enables multiplex SERS detection of tumor in vivo. As illustrated in Fig. 21a, a tumor model was surgically attached to a rat esophagus.263 In order to develop a tight seal between the rat esophagus and the tumor explants, a dermabond was applied by using a 20 μL pipette tip (Fig. 21b). The dermabond could be beneficial also in staining procedures, so that SERS NPs would not leak out from the esophagus (see the application principle in Fig. 21c). As depicted in Fig. 21d, the spectral-imaging probe was inserted into the guided tube, in order to image the antibody-conjugated SERS NPs that were attached on the luminal surface of the esophagus to target the cancer biomarkers EGFR and HER2 in vivo. The tumor location was visualized in vivo by ratiometric analyses between specific and non-specific binding.263
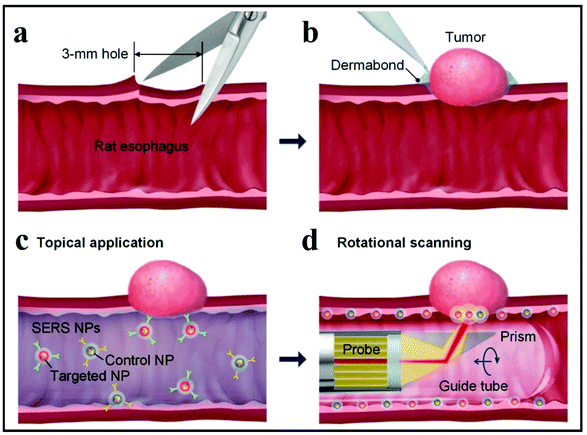 | ||
| Fig. 21 Schematic illustration of the development and endoscopic detection of an esophageal tumor model. (a) and (b) show the surgical bonding of tumor xenograft to the rat esophagus; (c) exhibits the SERS NP employment on the luminal surface for biomarker detection; and (d) shows the endoscopic imaging of the SERS signal from the imaging probe. Reproduced with permission.263 Copyright 2015, OSA. | ||
5. Future prospects and conclusion
To summarize, SERS activated platforms offer high sensitivity and promising multiplexing ability in bioanalysis and disease diagnosis (Fig. 22). The diversity of SERS-based assays, including label-based and label-free approaches on microfluidics chips, on paper-based substrates, etc., offers a flexible choice for their potential application in the biomedical and clinical fields. Moreover, by combining the SERS multiplex detection ability with in vivo endoscopic techniques, more promising applications arise as high-throughput assays and next-generation surgical treatments.SERS benefits from low cost, high sensitivity and rapid identification of the pathogen, which can guide a targeted treatment for the patient and therefore would save significant healthcare and financial resources. The intelligent analysis of spectral data would improve both the speed and the discrimination power of the technique. Several endeavors to employ big data analysis by using complex vector machines or in-house built discrimination algorithms for spectral data have been reported.162,166,186,187,264–266 The so-called fast diagnosis of sepsis is possible, according to research, by partially detecting via lateral flow immunoassay endogenous molecules produced only in the sepsis scenario and then identifying the pathogens by optical processes. The main advantage of this approach is the reduced time spent for bacterial classification, a real life-saving aspect in treating sepsis. An indispensable strategy for spectral pathology would be bringing Raman spectroscopy inside hospital premises and implementing biophotonics technologies in clinical routine procedures. However, recent advances in lasers, waveguide fiberoptics and microstructure-based optical fiber techniques267 enable in vivo clinical applications by their implementation in endoscopic setups.
Testing for the pathogen's existence in biological samples for infection diagnosis is employed nowadays in different fields of healthcare, and antibody-detecting rapid diagnostic tests for patients are envisioned in the near future.268 Lab-on-a-chip systems enable the incorporation of a range of analytic steps and therefore lead to multiplex testing and a fast throughput.
SERS microscopy can be very useful as a non-invasive, in situ fingerprinting technique that provides quantitative information due to the dependence of the optical response on concentration.269 However, SERS also holds biomolecular analysis benefits, as Raman spectra can probe different biomolecular components (including amino acids, nucleic acids, lipids, etc.) using quantifiable spectral deconvolution. For reference, spectral deconvolution has been applied for a long time in the pharmaceutical industries for quantitative analyses.270
SERS can detect single cells within biological samples. Such detection requires control of near-field nano-optic stability in the electromagnetic hotspots. It is also necessary to overcome the SERS signal fluctuations that limit reproducibility and/or to better understand the origins of the Raman signals from single cells. There are still areas of uncertainty in the SERS analysis of microorganisms; for instance, signal fluctuations are encountered that could be due to: (i) spontaneous variations in the conformation of the membrane's components, (ii) their adsorption state or photochemical effects in the vicinity of the metal surface, or (iii) heterogeneous membrane coverage with SERS-active substrates synthesized in situ. As the electromagnetic enhancement is the main contribution to the SERS signal, changes induced at the dielectric–metal interface, like controlled modification of refractive index, are highly important.
In conclusion, SERS benefits from the high sensitivity and the instrumental benefits of Raman spectroscopy (non-invasive analysis, minimal or no sample preparation, specificity down to small structural changes and high spatial resolution, portability, and miniaturization). SERS also has the ability to minimize spectral recording time, hence allowing the detection of a significant and statistically relevant number of spectra for valid multivariate analysis in a very short time. We believe that a wide SERS database comprising most common and clinically relevant pathogens would spur data analysis by generating statistically relevant input data for the development of new multi-level clustering algorithms.
Abbreviations
| AFM | Atomic force microscopy |
| AIs | Autoinducers |
| ALD | Atomic layer deposition |
| AMPs | Antimicrobial peptides |
| AMR | Antibiotic multiresistance |
| ASE | Amplified spontaneous emission |
| AST | Antibiotic susceptibility testing |
| ATP | Adenosine triphosphate |
| BOE | Buffered oxide etchant |
| CFUs | Colony-forming units |
| CM | Chemical enhancement |
| CTC | Circulating tumor cell |
| DCDR | Drop-coating deposition Raman |
| DEP | Dielectrophoresis |
| E. coli | Escherichia coli |
| EF | Enhancement factor |
| ELISA | Enzyme-linked immunosorbent assay |
| EM | Electromagnetic enhancement |
| EPS | Extracellular polymeric substances |
| ERS | Electronic Raman scattering |
| FDTD | Finite-difference time-domain |
| FHCC | Fuzzy hierarchical cross-clustering |
| GC–MS | Gas chromatography–mass spectrometry |
| GO | Graphene oxide |
| GI | Gastrointestinal |
| HTS | High-throughput screening |
| IFI | Invasive fungal infections |
| INRA | Immobilized nanorod assemblies |
| LBL | Layer-by-layer |
| LC–MS | Liquid chromatography–mass spectrometry |
| LDA | Linear discriminant analysis |
| LoC | Lab-on-a-chip |
| LSPs | Localized surface plasmons |
| LSPR | Localized metal surface plasmon resonance |
| MIC | Minimum inhibitory concentration |
| MIM | Metal–insulator–metal |
| MRS | Molecular Raman scattering |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MS | Molecular sentinel |
| NA | Numerical aperture |
| NBNS | Nanoplate–bacteria–nanorod supercrystals |
| NPs | Nanoparticles |
| nSTFs | Nano-sculptured thin films |
| OEG | Oligoethylene |
| PCA | Principal component analysis |
| PCR | Polymerase chain reaction |
| PDMS | Polydimethylsiloxane |
| PEI | Polyethylenimine |
| PET | Polyester |
| PLS | Partial least squares regression |
| PoC | Point-of-care |
| PS-SERES | Plasmon-sampled surface-enhanced Raman excitation spectroscopy |
| PU | Polyurethane |
| RMS | Raman microspectroscopy |
| RSD | Relative standard deviation |
| SAM | Self-assembled monolayer |
| SEM | Scanning electron microscopy |
| SERES | Surface-enhanced Raman excitation spectroscopy |
| SERS | Surface-enhanced Raman scattering |
| SORS | Spatial offset Raman scattering |
| SPR | Surface plasmon resonance |
| TDM | Therapeutic drug monitoring |
| TEM | Transmission electron microscopy |
| TLC | Thin-layer chromatography |
| UTAM | Ultrathin alumina mask |
| UTI | Urinary tract infection |
| VOC | Volatile organic compounds |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
N. E. Dina wishes to acknowledge a grant from the Ministry of Research, Innovation and Digitization, CNCS/CCCDI – UEFISCDI, project number PN-III-P1-1.1-PD-2016-0475 within PNCDI III and the financial support through the Core Program, Project No. PN 19 35 02 01. Liwu Zhang acknowledges support from the National Natural Science Foundation of China (No. 21677037 and 21976030) and the Natural Science Foundation of Shanghai (No. 17ZR1440200 and No. 19ZR1471200). V. K. V. acknowledges support from the Royal Society through the University Research Fellowships. V. K. V. and L. Z. acknowledge the International Collaboration Awards 2020 of the Royal Society (No. ICA\R1\201088).References
- K. Kneipp, Y. Wang, H. Kneipp, L. T. Perelman, I. Itzkan, R. R. Dasari and M. S. Feld, Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS), Phys. Rev. Lett., 1997, 78, 1667–1670 CrossRef CAS.
- S. Nie and S. R. Emory, Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- M. Fan and A. G. Brolo, Silver nanoparticles self assembly as SERS substrates with near single molecule detection limit, Phys. Chem. Chem. Phys., 2009, 11, 7381–7389 RSC.
- E. J. Blackie, E. C. Le Ru and P. G. Etchegoin, Single-Molecule Surface-Enhanced Raman Spectroscopy of Nonresonant Molecules, J. Am. Chem. Soc., 2009, 131, 14466–14472 CrossRef CAS PubMed.
- R. Tuma, Raman spectroscopy of proteins: from peptides to large assemblies, J. Raman Spectrosc., 2005, 36, 307–319 CrossRef CAS.
- A. Jaworska, S. Fornasaro, V. Sergo and A. Bonifacio, Potential of surface enhanced Raman spectroscopy (SERS) in therapeutic drug monitoring (TDM). A critical review, Biosensors, 2016, 6, 47 CrossRef PubMed.
- D. Cialla-May, X. S. Zheng, K. Weber and J. Popp, Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: from cells to clinics, Chem. Soc. Rev., 2017, 46, 3945–3961 RSC.
- E. Pyrak, J. Krajczewski, A. Kowalik, A. Kudelski and A. Jaworska, Surface Enhanced Raman Spectroscopy for DNA Biosensors-How Far Are We?, Molecules, 2019, 24, 4423 CrossRef CAS PubMed.
- M. Chisanga, H. Muhamadali, D. Ellis and R. Goodacre, Enhancing Disease Diagnosis: Biomedical Applications of Surface-Enhanced Raman Scattering, Appl. Sci., 2019, 9, 1163 CrossRef CAS.
- A. Locke, S. Fitzgerald and A. Mahadevan-Jansen, Advances in Optical Detection of Human-Associated Pathogenic Bacteria, Molecules, 2020, 25, 5256 CrossRef CAS PubMed.
- J. A. Dieringer, A. D. McFarland, N. C. Shah, D. A. Stuart, A. V. Whitney, C. R. Yonzon, M. A. Young, X. Zhang and R. P. Van Duyne, Introductory Lecture Surface enhanced Raman spectroscopy: new materials, concepts, characterization tools, and applications, Faraday Discuss., 2006, 132, 9–26 RSC.
- M. Moskovits, Surface-enhanced spectroscopy, Rev. Mod. Phys., 1985, 57, 783–826 CrossRef CAS.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. García de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Käll, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J.-F. Li, X. Y. Ling, S. A. Maier, T. Mayerhöfer, M. Moskovits, K. Murakoshi, J.-M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlücker, L.-L. Tay, K. G. Thomas, Z.-Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzán, Present and Future of Surface-Enhanced Raman Scattering, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- R. Aroca, Surface-enhanced vibrational spectroscopy, John Wiley & Sons, 2006 Search PubMed.
- J. Suh and M. Moskovits, Surface-enhanced Raman spectroscopy of amino acids and nucleotide bases adsorbed on silver, J. Am. Chem. Soc., 1986, 108, 4711–4718 CrossRef CAS.
- C. M. Muntean, N. E. Dina, M. Coroş, N. Toşa, A. I. Turza and M. Dan, Graphene/silver nanoparticles-based surface-enhanced Raman spectroscopy detection platforms: Application in the study of DNA molecules at low pH, J. Raman Spectrosc., 2019, 50, 1849–1860 CrossRef CAS.
- J. Granger, N. Schlotter, A. Crawford and M. Porter, Prospects for point-of-care pathogen diagnostics using surface-enhanced Raman scattering (SERS), Chem. Soc. Rev., 2016, 45, 3865–3882 RSC.
- T. J. Moore, A. S. Moody, T. D. Payne, G. M. Sarabia, A. R. Daniel and B. Sharma, In Vitro and In Vivo SERS Biosensing for Disease Diagnosis, Biosensors, 2018, 8, 46 CrossRef PubMed.
- Y. Liu, H. Zhou, Z. Hu, G. Yu, D. Yang and J. Zhao, Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review, Biosens. Bioelectron., 2017, 94, 131–140 CrossRef CAS PubMed.
- D. van Lierop, K. Faulds and D. Graham, Separation Free DNA Detection Using Surface Enhanced Raman Scattering, Anal. Chem., 2011, 83, 5817–5821 CrossRef CAS PubMed.
- D. van Lierop, Ž. Krpetić, L. Guerrini, I. A. Larmour, J. A. Dougan, K. Faulds and D. Graham, Positively charged silver nanoparticles and their effect on surface-enhanced Raman scattering of dye-labelled oligonucleotides, Chem. Commun., 2012, 48, 8192–8194 RSC.
- K. Gracie, M. Moores, W. E. Smith, K. Harding, M. Girolami, D. Graham and K. Faulds, Preferential Attachment of Specific Fluorescent Dyes and Dye Labeled DNA Sequences in a Surface Enhanced Raman Scattering Multiplex, Anal. Chem., 2016, 88, 1147–1153 CrossRef CAS PubMed.
- H. N. Wang, A. M. Fales and T. Vo-Dinh, Plasmonics-based SERS nanobiosensor for homogeneous nucleic acid detection, Nanomedicine, 2015, 11, 811–814 CrossRef CAS PubMed.
- L. Novotny and B. Hecht, in Principles of Nano-Optics, Cambridge University Press, Cambridge, 2006, pp. 378–418, DOI:10.1017/CBO9780511813535.013.
- V. Joseph, A. Matschulat, J. Polte, S. Rolf, F. Emmerling and J. Kneipp, SERS enhancement of gold nanospheres of defined size, J. Raman Spectrosc., 2011, 42, 1736–1742 CrossRef CAS.
- H. Yockell-Lelièvre, F. Lussier and J. F. Masson, Influence of the Particle Shape and Density of Self-Assembled Gold Nanoparticle Sensors on LSPR and SERS, J. Phys. Chem. C, 2015, 119, 28577–28585 CrossRef.
- Q. Jiwei, L. Yudong, Y. Ming, W. Qiang, C. Zongqiang, W. Wudeng, L. Wenqiang, Y. Xuanyi, X. Jingjun and S. Qian, Large-area high-performance SERS substrates with deep controllable sub-10-nm gap structure fabricated by depositing Au film on the cicada wing, Nanoscale Res. Lett., 2013, 8, 437 CrossRef PubMed.
- B. Sharma, M. Fernanda Cardinal, S. L. Kleinman, N. G. Greeneltch, R. R. Frontiera, M. G. Blaber, G. C. Schatz and R. P. Van Duyne, High-performance SERS substrates: Advances and challenges, MRS Bull., 2013, 38, 615–624 CrossRef CAS.
- L. Chen, L. Luo, Z. Chen, M. Zhang, J. A. Zapien, C. S. Lee and S. T. Lee, ZnO/Au Composite Nanoarrays As Substrates for Surface-Enhanced Raman Scattering Detection, J. Phys. Chem. C, 2010, 114, 93–100 CrossRef CAS.
- H. Huang, C. Shende, A. Sengupta, F. Inscore, C. Brouillette, W. Smith and S. Farquharson, Surface-enhanced Raman spectra of melamine and other chemicals using a 1550 nm (retina-safe) laser, J. Raman Spectrosc., 2012, 43, 701–705 CrossRef CAS.
- A. Campion and P. Kambhampati, Surface-enhanced Raman scattering, Chem. Soc. Rev., 1998, 27, 241–250 RSC.
- K. Kneipp, Y. Wang, R. R. Dasari, M. S. Feld, B. D. Gilbert, J. Janni and J. I. Steinfeld, Near-infrared surface-enhanced Raman scattering of trinitrotoluene on colloidal gold and silver, Spectrochim. Acta, Part A, 1995, 51, 2171–2175 CrossRef.
- J. I. Jerez-Rozo, O. M. Primera-Pedrozo, M. A. Barreto-Caban and S. P. Hernandez-Rivera, Enhanced Raman Scattering of 2,4,6-TNT Using Metallic Colloids, IEEE Sens. J., 2008, 8, 974–982 CAS.
- E. C. Le Ru, E. Blackie, M. Meyer and P. G. Etchegoin, Surface Enhanced Raman Scattering Enhancement Factors: A Comprehensive Study, J. Phys. Chem. C, 2007, 111, 13794–13803 CrossRef CAS.
- J. Jehlička, H. G. M. Edwards and A. Oren, Raman Spectroscopy of Microbial Pigments, Appl. Environ. Microbiol., 2014, 80, 3286–3295 CrossRef PubMed.
- A. C. Moţ, C. Bischin, G. Damian, A. A. A. Attia, E. Gal, N. E. Dina, N. Leopold and R. Silaghi-Dumitrescu, Fe(III) - Sulfide interaction in globins: Characterization and quest for a putative Fe(IV)-sulfide species, J. Inorg. Biochem., 2018, 179, 32–39 CrossRef PubMed.
- A. C. Moţ, M. Pârvu, A. E. Pârvu, O. Roşca-Casian, N. E. Dina, N. Leopold, R. Silaghi-Dumitrescu and C. Mircea, Reversible naftifine-induced carotenoid depigmentation in Rhodotorula mucilaginosa (A. Jörg.) F.C. Harrison causing onychomycosis, Sci. Rep., 2017, 7, 11125 CrossRef PubMed.
- G. Mei, N. Mamaeva, S. Ganapathy, P. Wang, W. J. DeGrip and K. J. Rothschild, Raman spectroscopy of a near infrared absorbing proteorhodopsin: Similarities to the bacteriorhodopsin O photointermediate, PLoS One, 2018, 13, e0209506 CrossRef CAS PubMed.
- K. Tian, W. Wang, X. Wang, S. Chen, J. Zhao, F. Tang, X. Nie and C. Han, Detection of flavin mononucleotide using silver nanorod array with simultaneous Raman scattering enhancement and fluorescence quenching, Opt. Eng., 2019, 58, 084109 CAS.
- N. E. Dina, A. Leş, A. Baricz, T. Szöke-Nagy, N. Leopold, C. Sârbu and H. L. Banciu, Discrimination of haloarchaeal genera using Raman spectroscopy and robust methods for multivariate data analysis, J. Raman Spectrosc., 2017, 48, 1122–1126 CrossRef CAS.
- B. Ma, P. Li, L. Yang and J. Liu, Based on time and spatial-resolved SERS mapping strategies for detection of pesticides, Talanta, 2015, 141, 1–7 CrossRef CAS PubMed.
- C. Ortiz, D. Zhang, Y. Xie, A. E. Ribbe and D. Ben-Amotz, Validation of the drop coating deposition Raman method for protein analysis, Anal. Biochem., 2006, 353, 157–166 CrossRef CAS PubMed.
- J. Filik and N. Stone, Drop coating deposition Raman spectroscopy of protein mixtures, Analyst, 2007, 132, 544–550 RSC.
- A. Ştefancu, V. Moişoiu, R. Couti, I. Andras, R. Rahota, D. Crişan, I. E. Pavel, C. Socaciu, N. Leopold and N. Crişan, Combining SERS analysis of serum with PSA levels for improving the detection of prostate cancer, Nanomedicine, 2018, 13, 2455–2467 CrossRef PubMed.
- N. C. Dingari, G. L. Horowitz, J. W. Kang, R. R. Dasari and I. Barman, Raman Spectroscopy Provides a Powerful Diagnostic Tool for Accurate Determination of Albumin Glycation, PLoS One, 2012, 7, e32406 CrossRef CAS PubMed.
- A. Ştefancu, V. Moişoiu, C. Bocşa, Z. Bálint, D.-T. Cosma, I. A. Veresiu, V. Chiş, N. Leopold and F. Elec, SERS-based quantification of albuminuria in the normal-to-mildly increased range, Analyst, 2018, 143, 5372–5379 RSC.
- L. Chen, N. Mungroo, L. Daikuara and S. Neethirajan, Label-free NIR-SERS discrimination and detection of foodborne bacteria by in situ synthesis of Ag colloids, J. Nanobiotechnol., 2015, 13, 45 CrossRef PubMed.
- N. E. Dina, A. M. R. Gherman, V. Chiş, C. Sârbu, A. Wieser, D. Bauer and C. Haisch, Characterization of Clinically Relevant Fungi via SERS Fingerprinting Assisted by Novel Chemometric Models, Anal. Chem., 2018, 90, 2484–2492 CrossRef CAS PubMed.
- H. Zhou, D. Yang, N. P. Ivleva, N. E. Mircescu, R. Niessner and C. Haisch, SERS Detection of Bacteria in Water by in Situ Coating with Ag Nanoparticles, Anal. Chem., 2014, 86, 1525–1533 CrossRef CAS PubMed.
- D.-R. Hermann, D. Lilek, C. Daffert, I. Fritz, S. Weinberger, V. Rumpler, B. Herbinger and K. Prohaska, In situ based surface-enhanced Raman spectroscopy (SERS) for the fast and reproducible identification of PHB producers in cyanobacterial cultures, Analyst, 2020, 145, 5242–5251 RSC.
- X. Wu, Y.-W. Huang, B. Park, R. A. Tripp and Y. Zhao, Differentiation and classification of bacteria using vancomycin functionalized silver nanorods array based surface-enhanced Raman spectroscopy and chemometric analysis, Talanta, 2015, 139, 96–103 CrossRef CAS PubMed.
- N. E. Dina, H. Zhou, A. Colnita, N. Leopold, T. Szoke-Nagy, C. Coman and C. Haisch, Rapid single-cell detection and identification of pathogens by using surface-enhanced Raman spectroscopy, Analyst, 2017, 142, 1782–1789 RSC.
- S. Nie and S. R. Emory, Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- A. R. M. N. Afrooz, S. T. Sivalapalan, C. J. Murphy, S. M. Hussain, J. J. Schlager and N. B. Saleh, Spheres vs. rods: The shape of gold nanoparticles influences aggregation and deposition behavior, Chemosphere, 2013, 91, 93–98 CrossRef CAS PubMed.
- V. G. Pol, A. Gedanken and J. Calderon-Moreno, Deposition of Gold Nanoparticles on Silica Spheres: A Sonochemical Approach, Chem. Mater., 2003, 15, 1111–1118 CrossRef CAS.
- K. Kucharczyk, J. D. Rybka, M. Hilgendorff, M. Krupinski, M. Slachcinski, A. Mackiewicz, M. Giersig and H. Dams-Kozlowska, Composite spheres made of bioengineered spider silk and iron oxide nanoparticles for theranostics applications, PLoS One, 2019, 14, e0219790 CrossRef CAS PubMed.
- C. Farcău and S. Aştilean, Mapping the SERS Efficiency and Hot-Spots Localization on Gold Film over Nanospheres Substrates, J. Phys. Chem. C, 2010, 114, 11717–11722 CrossRef.
- X. Wang, X. Bai, Z. Pang, J. Zhu, Y. Wu, H. Yang, Y. Qi and X. Wen, Surface-enhanced Raman scattering by composite structure of gold nanocube-PMMA-gold film, Opt. Mater. Express, 2019, 9, 1872–1881 CrossRef CAS.
- L. Zhang, W. Zhang, F. Lu, Z. Yang, T. Xue, M. Liu, C. Meng, P. Li, D. Mao, T. Mei and J. Zhao, Azimuthal vector beam exciting silver triangular nanoprisms for increasing the performance of surface-enhanced Raman spectroscopy, Photonics Res., 2019, 7, 1447–1453 CrossRef CAS.
- L. Lu, A. Kobayashi, K. Tawa and Y. Ozaki, Silver Nanoplates with Special Shapes: Controlled Synthesis and Their Surface Plasmon Resonance and Surface-Enhanced Raman Scattering Properties, Chem. Mater., 2006, 18, 4894–4901 CrossRef CAS.
- Z. Fan, D. Senapati, S. A. Khan, A. K. Singh, A. Hamme, B. Yust, D. Sardar and P. C. Ray, Popcorn-shaped magnetic core-plasmonic shell multifunctional nanoparticles for the targeted magnetic separation and enrichment, label-free SERS imaging, and photothermal destruction of multidrug-resistant bacteria, Chemistry, 2013, 19, 2839–2847 CrossRef CAS PubMed.
- V. V. R. Sai, D. Gangadean, I. Niraula, J. M. F. Jabal, G. Corti, D. N. McIlroy, D. Eric Aston, J. R. Branen and P. J. Hrdlicka, Silica Nanosprings Coated with Noble Metal Nanoparticles: Highly Active SERS Substrates, J. Phys. Chem. C, 2011, 115, 453–459 CrossRef CAS.
- N. Zhao, Y. Wei, N. Sun, Q. Chen, J. Bai, L. Zhou, Y. Qin, M. Li and L. Qi, Controlled Synthesis of Gold Nanobelts and Nanocombs in Aqueous Mixed Surfactant Solutions, Langmuir, 2008, 24, 991–998 CrossRef CAS PubMed.
- A. Guerrero-Martínez, S. Barbosa, I. Pastoriza-Santos and L. M. Liz-Marzán, Nanostars shine bright for you: Colloidal synthesis, properties and applications of branched metallic nanoparticles, Curr. Opin. Colloid Interface Sci., 2011, 16, 118–127 CrossRef.
- S. Boca, D. Rugina, A. Pintea, L. Barbu-Tudoran and S. Astilean, Flower-shaped gold nanoparticles: synthesis, characterization and their application as SERS-active tags inside living cells, Nanotechnology, 2010, 22, 055702 CrossRef PubMed.
- M. Fernanda Cardinal, B. Rodríguez-González, R. A. Alvarez-Puebla, J. Pérez-Juste and L. M. Liz-Marzán, Modulation of Localized Surface Plasmons and SERS Response in Gold Dumbbells through Silver Coating, J. Phys. Chem. C, 2010, 114, 10417–10423 CrossRef CAS.
- Y. W. Lee and T. H. Shin, Synthesis of size-controlled urchin AgNPs and surface-enhanced Raman spectroscopy, J. Korean Inst. Electr. Electron. Mater. Eng., 2019, 32, 454–457 Search PubMed.
- C. P. Shaw, M. Fan, C. Lane, G. Barry, A. I. Jirasek and A. G. Brolo, Statistical Correlation Between SERS Intensity and Nanoparticle Cluster Size, J. Phys. Chem. C, 2013, 117, 16596–16605 CrossRef CAS.
- A. Lee, G. F. S. Andrade, A. Ahmed, M. L. Souza, N. Coombs, E. Tumarkin, K. Liu, R. Gordon, A. G. Brolo and E. Kumacheva, Probing Dynamic Generation of Hot-Spots in Self-Assembled Chains of Gold Nanorods by Surface-Enhanced Raman Scattering, J. Am. Chem. Soc., 2011, 133, 7563–7570 CrossRef CAS PubMed.
- A. Fargašová, R. Prucek, V. Ranc, A. Panáček, L. Kvítek and R. Zbořil, Influence of various chloride ion concentrations on silver nanoparticle transformations and effectiveness in surface enhanced Raman scattering for different excitation wavelengths, RSC Adv., 2015, 5, 9737–9744 RSC.
- J. Yoo, H. So, M. Yang and K.-J. Lee, Effect of chloride ion on synthesis of silver nanoparticle using retrieved silver chloride as a precursor from the electronic scrap, Appl. Surf. Sci., 2019, 475, 781–784 CrossRef CAS.
- L.-J. Xu, C. Zong, X.-S. Zheng, P. Hu, J.-M. Feng and B. Ren, Label-Free Detection of Native Proteins by Surface-Enhanced Raman Spectroscopy Using Iodide-Modified Nanoparticles, Anal. Chem., 2014, 86, 2238–2245 CrossRef CAS PubMed.
- M. Fan and A. G. Brolo, Self-assembled Au nanoparticles as substrates for surface-enhanced vibrational spectroscopy: optimization and electrochemical stability, ChemPhysChem, 2008, 9, 1899–1907 CrossRef CAS PubMed.
- K. L. Wustholz, A.-I. Henry, J. M. McMahon, R. G. Freeman, N. Valley, M. E. Piotti, M. J. Natan, G. C. Schatz and R. P. Van Duyne, Structure–Activity Relationships in Gold Nanoparticle Dimers and Trimers for Surface-Enhanced Raman Spectroscopy, J. Am. Chem. Soc., 2010, 132, 10903–10910 CrossRef CAS PubMed.
- L. O. Herrmann, V. K. Valev, C. Tserkezis, J. S. Barnard, S. Kasera, O. A. Scherman, J. Aizpurua and J. J. Baumberg, Threading plasmonic nanoparticle strings with light, Nat. Commun., 2014, 5, 4568 CrossRef CAS PubMed.
- T. Ding, V. K. Valev, A. R. Salmon, C. J. Forman, S. K. Smoukov, O. A. Scherman, D. Frenkel and J. J. Baumberg, Light-induced actuating nanotransducers, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 5503–5507 CrossRef CAS PubMed.
- H. Xu, J. Aizpurua, M. Kall and P. Apell, Electromagnetic contributions to single-molecule sensitivity in surface-enhanced raman scattering, Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 2000, 62, 4318–4324 CrossRef CAS PubMed.
- P. Tan, H. Li, J. Wang and S. C. B. Gopinath, Silver nanoparticle in biosensor and bioimaging: Clinical perspectives, Biotechnol. Appl. Biochem., 2021 DOI:10.1002/bab.2045.
- E. Babich, A. Redkov, I. Reduto and A. Lipovskii, Self-Assembled Silver–Gold Nanoisland Films on Glass for SERS Applications, Phys. Status Solidi RRL, 2018, 12, 1700226 CrossRef.
- S. Saha, M. Ghosh, B. Dutta and J. Chowdhury, Silver coated gold nanocolloids entrapped in organized Langmuir–Blodgett Film of stearic acid: Potential evidence of a new SERS active substrate, Appl. Surf. Sci., 2016, 362, 364–373 CrossRef CAS.
- K. Yuan, Q. Mei, X. Guo, Y. Xu, D. Yang, B. J. Sánchez, B. Sheng, C. Liu, Z. Hu, G. Yu, H. Ma, H. Gao, C. Haisch, R. Niessner, Z. Jiang and H. Zhou, Antimicrobial peptide based magnetic recognition elements and Au@Ag-GO SERS tags with stable internal standards: a three in one biosensor for isolation, discrimination and killing of multiple bacteria in whole blood, Chem. Sci., 2018, 9, 8781–8795 RSC.
- T. Kim, C.-H. Lee, S.-W. Joo and K. Lee, Kinetics of gold nanoparticle aggregation: experiments and modeling, J. Colloid Interface Sci., 2008, 318, 238–243 CrossRef CAS PubMed.
- S. Kundu and H. Liang, Polyelectrolyte-mediated non-micellar synthesis of monodispersed ‘aggregates’ of gold nanoparticles using a microwave approach, Colloids Surf., A, 2008, 330, 143–150 CrossRef CAS.
- N. Leopold, V. Chiş, N. E. Mircescu, O. T. Marişca, O. M. Buja, L. F. Leopold, C. Socaciu, C. Braicu, A. Irimie and I. Berindan-Neagoe, One step synthesis of SERS active colloidal gold nanoparticles by reduction with polyethylene glycol, Colloids Surf., A, 2013, 436, 133–138 CrossRef CAS.
- O. T. Marisca, K. Kantner, C. Pfeiffer, Q. Zhang, B. Pelaz, N. Leopold, W. J. Parak and J. Rejman, Comparison of the in Vitro Uptake and Toxicity of Collagen- and Synthetic Polymer-Coated Gold Nanoparticles, Nanomaterials, 2015, 5, 1418–1430 CrossRef CAS PubMed.
- L. F. Leopold, I. S. Tódor, Z. Diaconeasa, D. Rugină, A. Ştefancu, N. Leopold and C. Coman, Assessment of PEG and BSA-PEG gold nanoparticles cellular interaction, Colloids Surf., A, 2017, 532, 70–76 CrossRef CAS.
- N. G. Greeneltch, M. G. Blaber, A. I. Henry, G. C. Schatz and R. P. Van Duyne, Immobilized nanorod assemblies: fabrication and understanding of large area surface-enhanced Raman spectroscopy substrates, Anal. Chem., 2013, 85, 2297–2303 CrossRef CAS PubMed.
- H. T. Ngo, H.-N. Wang, A. M. Fales and T. Vo-Dinh, Label-free DNA biosensor based on SERS Molecular Sentinel on Nanowave chip, Anal. Chem., 2013, 85, 6378–6383 CrossRef CAS PubMed.
- H. Ngo, H.-N. Wang, B. Crawford, A. Fales and T. Vo-Dinh, Plasmonic SERS nanochips and nanoprobes for medical diagnostics and bio-energy applications, SPIE, 2017 Search PubMed.
- C. G. Khoury and T. Vo-Dinh, Plasmonic Nanowave Substrates for SERS: Fabrication and Numerical Analysis, J. Phys. Chem. C, 2012, 116, 7534–7545 CrossRef CAS PubMed.
- T. Vo-Dinh, M. Y. K. Hiromoto, G. M. Begun and R. L. Moody, Surface-enhanced Raman spectrometry for trace organic analysis, Anal. Chem., 1984, 56, 1667–1670 CrossRef CAS.
- A. Garg, W. Nam and W. Zhou, Reusable Surface-Enhanced Raman Spectroscopy Membranes and Textiles via Template-Assisted Self-Assembly and Micro/Nanoimprinting, ACS Appl. Mater. Interfaces, 2020, 12, 56290–56299 CrossRef CAS PubMed.
- P. Kubryk, R. Niessner and N. P. Ivleva, The origin of the band at around 730 cm−1 in the SERS spectra of bacteria: a stable isotope approach, Analyst, 2016, 141, 2874–2878 RSC.
- W. R. Premasiri, J. C. Lee, A. Sauer-Budge, R. Theberge, C. E. Costello and L. D. Ziegler, The biochemical origins of the surface-enhanced Raman spectra of bacteria: a metabolomics profiling by SERS, Anal. Bioanal. Chem., 2016, 408, 4631–4647 CrossRef CAS PubMed.
- D. Zhang, Y. Xie, S. K. Deb, V. J. Davison and D. Ben-Amotz, Isotope edited internal standard method for quantitative surface-enhanced Raman spectroscopy, Anal. Chem., 2005, 77, 3563–3569 CrossRef CAS PubMed.
- S. K. Deb, B. Davis, G. M. Knudsen, R. Gudihal, D. Ben-Amotz and V. J. Davisson, Detection and relative quantification of proteins by surface enhanced Raman using isotopic labels, J. Am. Chem. Soc., 2008, 130, 9624–9625 CrossRef CAS PubMed.
- J. J. Baumberg, T. A. Kelf, Y. Sugawara, S. Cintra, M. E. Abdelsalam, P. N. Bartlett and A. E. Russell, Angle-resolved surface-enhanced Raman scattering on metallic nanostructured plasmonic crystals, Nano Lett., 2005, 5, 2262–2267 CrossRef CAS PubMed.
- J. F. Li, Y. F. Huang, Y. Ding, Z. L. Yang, S. B. Li, X. S. Zhou, F. R. Fan, W. Zhang, Z. Y. Zhou and B. Ren, Shell-isolated nanoparticle-enhanced Raman spectroscopy, Nature, 2010, 464, 392–395 CrossRef CAS PubMed.
- Y. Wang, B. Yan and L. Chen, SERS tags: novel optical nanoprobes for bioanalysis, Chem. Rev., 2013, 113, 1391–1428 CrossRef CAS PubMed.
- H. Abramczyk and B. Brozek-Pluska, Raman imaging in biochemical and biomedical applications. Diagnosis and treatment of breast cancer, Chem. Rev., 2013, 113, 5766–5781 CrossRef CAS PubMed.
- J. Fang, S. Du, S. Lebedkin, Z. Li, R. Kruk, M. Kappes and H. Hahn, Gold mesostructures with tailored surface topography and their self-assembly arrays for surface-enhanced Raman spectroscopy, Nano Lett., 2010, 10, 5006–5013 CrossRef CAS PubMed.
- R. Zhang, Y. Zhang, Z. Dong, S. Jiang, C. Zhang, L. Chen, L. Zhang, Y. Liao, J. Aizpurua and Y. E. Luo, Chemical mapping of a single molecule by plasmon-enhanced Raman scattering, Nature, 2013, 498, 82–86 CrossRef CAS PubMed.
- D.-K. Lim, K.-S. Jeon, H. M. Kim, J.-M. Nam and Y. D. Suh, Nanogap-engineerable Raman-active nanodumbbells for single-molecule detection, Nat. Mater., 2010, 9, 60–67 CrossRef CAS PubMed.
- D.-K. Lim, K.-S. Jeon, J.-H. Hwang, H. Kim, S. Kwon, Y. D. Suh and J.-M. Nam, Highly uniform and reproducible surface-enhanced Raman scattering from DNA-tailorable nanoparticles with 1 nm interior gap, Nat. Nanotechnol., 2011, 6, 452–460 CrossRef CAS PubMed.
- J. P. Camden, J. A. Dieringer, Y. Wang, D. J. Masiello, L. D. Marks, G. C. Schatz and R. P. Van Duyne, Probing the structure of single-molecule surface-enhanced Raman scattering hot spots, J. Am. Chem. Soc., 2008, 130, 12616–12617 CrossRef CAS PubMed.
- E. D. Diebold, N. H. Mack, S. K. Doorn and E. Mazur, Femtosecond laser-nanostructured substrates for surface-enhanced Raman scattering, Langmuir, 2009, 25, 1790–1794 CrossRef CAS PubMed.
- X. Zhang, Y. Zheng, X. Liu, W. Lu, J. Dai, D. Y. Lei and D. R. MacFarlane, Hierarchical porous plasmonic metamaterials for reproducible ultrasensitive surface–enhanced Raman spectroscopy, Adv. Mater., 2015, 27, 1090–1096 CrossRef CAS PubMed.
- M. Hu, F. S. Ou, W. Wu, I. Naumov, X. Li, A. M. Bratkovsky, R. S. Williams and Z. Li, Gold nanofingers for molecule trapping and detection, J. Am. Chem. Soc., 2010, 132, 12820–12822 CrossRef CAS PubMed.
- Q. Fu, Z. Zhan, J. Dou, X. Zheng, R. Xu, M. Wu and Y. Lei, Highly reproducible and sensitive SERS substrates with Ag inter-nanoparticle gaps of 5 nm fabricated by ultrathin aluminum mask technique, ACS Appl. Mater. Interfaces, 2015, 7, 13322–13328 CrossRef CAS PubMed.
- H. T. Miyazaki and Y. Kurokawa, Squeezing visible light waves into a 3 nm-thick and 55 nm-long plasmon cavity, Phys. Rev. Lett., 2006, 96, 097401 CrossRef PubMed.
- D. R. Ward, F. Hüser, F. Pauly, J. C. Cuevas and D. Natelson, Optical rectification and field enhancement in a plasmonic nanogap, Nat. Nanotechnol., 2010, 5, 732–736 CrossRef CAS PubMed.
- T. W. Ebbesen, H. J. Lezec, H. Ghaemi, T. Thio and P. A. Wolff, Extraordinary optical transmission through sub-wavelength hole arrays, Nature, 1998, 391, 667–669 CrossRef CAS.
- W. L. Barnes, A. Dereux and T. W. Ebbesen, Surface plasmon subwavelength optics, Nature, 2003, 424, 824–830 CrossRef CAS PubMed.
- L. Novotny and N. Van Hulst, Antennas for light, Nat. Photonics, 2011, 5, 83–90 CrossRef CAS.
- M. Seo, H. Park, S. Koo, D. Park, J. Kang, O. Suwal, S. Choi, P. Planken, G. Park and N. Park, Terahertz field enhancement by a metallic nano slit operating beyond the skin-depth limit, Nat. Photonics, 2009, 3, 152–156 CrossRef CAS.
- N. J. Halas, S. Lal, W.-S. Chang, S. Link and P. Nordlander, Plasmons in strongly coupled metallic nanostructures, Chem. Rev., 2011, 111, 3913–3961 CrossRef CAS PubMed.
- A. Fursina, S. Lee, R. Sofin, I. Shvets and D. Natelson, Nanogaps with very large aspect ratios for electrical measurements, Appl. Phys. Lett., 2008, 92, 113102 CrossRef.
- D. R. Ward, N. K. Grady, C. S. Levin, N. J. Halas, Y. Wu, P. Nordlander and D. Natelson, Electromigrated nanoscale gaps for surface-enhanced Raman spectroscopy, Nano Lett., 2007, 7, 1396–1400 CrossRef CAS PubMed.
- K. J. Savage, M. M. Hawkeye, R. Esteban, A. G. Borisov, J. Aizpurua and J. J. Baumberg, Revealing the quantum regime in tunnelling plasmonics, Nature, 2012, 491, 574–577 CrossRef CAS PubMed.
- X. Chen, H.-R. Park, M. Pelton, X. Piao, N. C. Lindquist, H. Im, Y. J. Kim, J. S. Ahn, K. J. Ahn, N. Park, D.-S. Kim and S.-H. Oh, Atomic layer lithography of wafer-scale nanogap arrays for extreme confinement of electromagnetic waves, Nat. Commun., 2013, 4, 2361 CrossRef PubMed.
- J. Song, W. Nam and W. Zhou, Scalable high–performance nanolaminated SERS substrates based on multistack vertically oriented plasmonic nanogaps, Adv. Mater. Technol., 2019, 4, 1800689 CrossRef.
- W. Nam, X. Ren, S. A. S. Tali, P. Ghassemi, I. Kim, M. Agah and W. Zhou, Refractive-index-insensitive nanolaminated SERS substrates for label-free Raman profiling and classification of living cancer cells, Nano Lett., 2019, 19, 7273–7281 CrossRef CAS PubMed.
- A. Lorén, J. Engelbrektsson, C. Eliasson, M. Josefson, J. Abrahamsson, M. Johansson and K. Abrahamsson, Internal standard in surface-enhanced Raman spectroscopy, Anal. Chem., 2004, 76, 7391–7395 CrossRef PubMed.
- H.-Y. Chen, M.-H. Lin, C.-Y. Wang, Y.-M. Chang and S. Gwo, Large-scale hot spot engineering for quantitative SERS at the single-molecule scale, J. Am. Chem. Soc., 2015, 137, 13698–13705 CrossRef CAS PubMed.
- C.-F. Chen, S.-D. Tzeng, H.-Y. Chen, K.-J. Lin and S. Gwo, Tunable plasmonic response from alkanethiolate-stabilized gold nanoparticle superlattices: evidence of near-field coupling, J. Am. Chem. Soc., 2008, 130, 824–826 CrossRef CAS PubMed.
- Y. Zhou, R. Ding, P. Joshi and P. Zhang, Quantitative surface-enhanced Raman measurements with embedded internal reference, Anal. Chim. Acta, 2015, 874, 49–53 CrossRef CAS PubMed.
- W. Shen, X. Lin, C. Jiang, C. Li, H. Lin, J. Huang, S. Wang, G. Liu, X. Yan and Q. Zhong, Reliable quantitative SERS analysis facilitated by core–shell nanoparticles with embedded internal standards, Angew. Chem., Int. Ed., 2015, 54, 7308–7312 CrossRef CAS PubMed.
- N. E. Mircescu, M. Oltean, V. Chiş and N. Leopold, FTIR, FT-Raman, SERS and DFT study on melamine, Vib. Spectrosc., 2012, 62, 165–171 CrossRef CAS.
- A. Beganović, T. Nuener, F. Meischl, S. E. Stuppner, M. Rainer, G. K. Bonn and C. W. Huck, Quantification of melamine in infant formula using a handheld Raman spectrometer–Performance boost with customized Arduino-controlled rotation setup, Talanta, 2020, 209, 120488 CrossRef PubMed.
- H. Wei, A. McCarthy, J. Song, W. Zhou and P. J. Vikesland, Quantitative SERS by hot spot normalization–surface enhanced Rayleigh band intensity as an alternative evaluation parameter for SERS substrate performance, Faraday Discuss., 2017, 205, 491–504 RSC.
- H. Wei, W. Leng, J. Song, M. R. Willner, L. C. Marr, W. Zhou and P. J. Vikesland, Improved quantitative SERS enabled by surface plasmon enhanced elastic light scattering, Anal. Chem., 2018, 90, 3227–3237 CrossRef CAS PubMed.
- W. Nam, Y. Zhao, J. Song, S. Ali Safiabadi Tali, S. Kang, W. Zhu, H. J. Lezec, A. Agrawal, P. J. Vikesland and W. Zhou, Plasmonic Electronic Raman Scattering as Internal Standard for Spatial and Temporal Calibration in Quantitative Surface-Enhanced Raman Spectroscopy, J. Phys. Chem. Lett., 2020, 11, 9543–9551 CrossRef CAS PubMed.
- W. Nam, X. Ren, I. Kim, J. Strobl, M. Agah and W. Zhou, Plasmonically Calibrated Label-Free Surface-Enhanced Raman Spectroscopy for Improved Multivariate Analysis of Living Cells in Cancer Subtyping and Drug Testing, Anal. Chem., 2021, 93, 4601–4610 CrossRef CAS PubMed.
- S. Kumar, R. Gopinathan, G. K. Chandra, S. Umapathy and D. K. Saini, Rapid detection of bacterial infection and viability assessment with high specificity and sensitivity using Raman microspectroscopy, Anal. Bioanal. Chem., 2020, 412, 2505–2516 CrossRef CAS PubMed.
- C. Fleischmann, D. O. Thomas-Rueddel, M. Hartmann, C. S. Hartog, T. Welte, S. Heublein, U. Dennler and K. Reinhart, Hospital Incidence and Mortality Rates of Sepsis: An Analysis of Hospital Episode (DRG) Statistics in Germany From 2007 to 2013, Dtsch. Arzteblatt Int., 2016, 113, 159–166 Search PubMed.
- WHO, World Health Statistics 2015, 2016 Search PubMed.
- E. M. Cody and B. P. Dixon, Hemolytic Uremic Syndrome, Pediatr. Clin. North Am., 2019, 66, 235–246 CrossRef PubMed.
- J. Teng, F. Yuan, Y. Ye, L. Zheng, L. Yao, F. Xue, W. Chen and B. Li, Aptamer-Based Technologies in Foodborne Pathogen Detection, Front. Microbiol., 2016, 7, 1426 Search PubMed.
- S. M. Yoo and S. Y. Lee, Optical Biosensors for the Detection of Pathogenic Microorganisms, Trends Biotechnol., 2016, 34, 7–25 CrossRef CAS PubMed.
- S. S. Zhao, N. Bukar, J. L. Toulouse, D. Pelechacz, R. Robitaille, J. N. Pelletier and J. F. Masson, Miniature multi-channel SPR instrument for methotrexate monitoring in clinical samples, Biosens. Bioelectron., 2015, 64, 664–670 CrossRef CAS PubMed.
- Y. Liu, Q. Liu, S. Chen, F. Cheng, H. Wang and W. Peng, Surface Plasmon Resonance Biosensor Based on Smart Phone Platforms, Sci. Rep., 2015, 5, 12864 CrossRef CAS PubMed.
- K. Kneipp, A. S. Haka, H. Kneipp, K. Badizadegan, N. Yoshizawa, C. Boone, K. E. Shafer-Peltier, J. T. Motz, R. R. Dasari and M. S. Feld, Surface-Enhanced Raman Spectroscopy in Single Living Cells Using Gold Nanoparticles, Appl. Spectrosc., 2002, 56, 150–154 CrossRef CAS.
- P. Wang, S. Pang, J. Chen, L. McLandsborough, S. R. Nugen, M. Fan and L. He, Label-free mapping of single bacterial cells using surface-enhanced Raman spectroscopy, Analyst, 2016, 141, 1356–1362 RSC.
- B. Hecht and L. Novotny, in Principles of Nano-Optics, Cambridge University Press, Cambridge, 2006, pp. 1–12, DOI:10.1017/CBO9780511813535.002.
- L. E. Jamieson and H. J. Byrne, Vibrational spectroscopy as a tool for studying drug-cell interaction: Could high throughput vibrational spectroscopic screening improve drug development?, Vib. Spectrosc., 2017, 91, 16–30 CrossRef CAS.
- I. F. Cheng, H.-C. Chang, T.-Y. Chen, C. Hu and F.-L. Yang, Rapid (<5 min) Identification of Pathogen in Human Blood by Electrokinetic Concentration and Surface-Enhanced Raman Spectroscopy, Sci. Rep., 2013, 3, 1–8 Search PubMed.
- H. Muhamadali, A. Subaihi, M. Mohammadtaheri, Y. Xu, D. I. Ellis, R. Ramanathan, V. Bansal and R. Goodacre, Rapid, accurate, and comparative differentiation of clinically and industrially relevant microorganisms via multiple vibrational spectroscopic fingerprinting, Analyst, 2016, 141, 5127–5136 RSC.
- O. Gorelik, N. Levy, L. Shaulov, K. Yegodayev, M. M. Meijler and N. Sal-Man, Vibrio cholerae autoinducer-1 enhances the virulence of enteropathogenic Escherichia coli, Sci. Rep., 2019, 9, 4122 CrossRef PubMed.
- D. G. Davies, M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton and E. P. Greenberg, The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm, Science, 1998, 280, 295–298 CrossRef CAS PubMed.
- D. H. Lenz, K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen and B. L. Bassler, The Small RNA Chaperone Hfq and Multiple Small RNAs Control Quorum Sensing in Vibrio harveyi and Vibrio cholerae, Cell, 2004, 118, 69–82 CrossRef CAS PubMed.
- L. Passador, J. Cook, M. Gambello, L. Rust and B. Iglewski, Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication, Science, 1993, 260, 1127–1130 CrossRef CAS PubMed.
- N. P. Ivleva, M. Wagner, H. Horn, R. Niessner and C. Haisch, Towards a nondestructive chemical characterization of biofilm matrix by Raman microscopy, Anal. Bioanal. Chem., 2009, 393, 197–206 CrossRef CAS PubMed.
- Y. Li, C. Lu, S. Zhou, M.-L. Fauconnier, F. Gao, B. Fan, J. Lin, F. Wang and J. Zheng, Sensitive and simultaneous detection of different pathogens by surface-enhanced Raman scattering based on aptamer and Raman reporter co-mediated gold tags, Sens. Actuators, B, 2020, 317, 128182 CrossRef CAS.
- Y. Pang, N. Wan, L. Shi, C. Wang, Z. Sun, R. Xiao and S. Wang, Dual-recognition surface-enhanced Raman scattering(SERS)biosensor for pathogenic bacteria detection by using vancomycin-SERS tags and aptamer-Fe3O4@Au, Anal. Chim. Acta, 2019, 1077, 288–296 CrossRef CAS PubMed.
- C. Wang, J. Wang, M. Li, X. Qu, K. Zhang, Z. Rong, R. Xiao and S. Wang, A rapid SERS method for label-free bacteria detection using polyethylenimine-modified Au-coated magnetic microspheres and Au@Ag nanoparticles, Analyst, 2016, 141, 6226–6238 RSC.
- M. A. Tahir, X. Zhang, H. Cheng, D. Xu, Y. Feng, G. Sui, H. Fu, V. K. Valev, L. Zhang and J. Chen, Klarite as a label-free SERS-based assay: a promising approach for atmospheric bioaerosol detection, Analyst, 2020, 145, 277–285 RSC.
- H. Zhou, D. Yang, N. P. Ivleva, N. E. Mircescu, S. Schubert, R. Niessner, A. Wieser and C. Haisch, Label-Free in Situ Discrimination of Live and Dead Bacteria by Surface-Enhanced Raman Scattering, Anal. Chem., 2015, 87, 6553–6561 CrossRef CAS PubMed.
- O. Prakash, S. Sil, T. Verma and S. Umapathy, Direct Detection of Bacteria Using Positively Charged Ag/Au Bimetallic Nanoparticles: A Label-free Surface-Enhanced Raman Scattering Study Coupled with Multivariate Analysis, J. Phys. Chem. C, 2020, 124, 861–869 CrossRef CAS.
- M. De, P. S. Ghosh and V. M. Rotello, Applications of Nanoparticles in Biology, Adv. Mater., 2008, 20, 4225–4241 CrossRef CAS.
- S. Kloß, B. Kampe, S. Sachse, P. Rösch, E. Straube, W. Pfister, M. Kiehntopf and J. Popp, Culture Independent Raman Spectroscopic Identification of Urinary Tract Infection Pathogens: A Proof of Principle Study, Anal. Chem., 2013, 85, 9610–9616 CrossRef PubMed.
- U. Neugebauer, S. Kloß, U.-C. Schröder, P. Rösch and J. Popp, Fast and Selective against bacteria culture-independent identification of pathogens from the urinary tract, Opt. Photonik, 2013, 8, 36–39 CrossRef CAS.
- U.-C. Schröder, C. Beleites, C. Assmann, U. Glaser, U. Hübner, W. Pfister, W. Fritzsche, J. Popp and U. Neugebauer, Detection of vancomycin resistances in enterococci within 3 ½ hours, Sci. Rep., 2015, 5, 1–7 Search PubMed.
- T. Y. Liu, Y. Chen, H. H. Wang, Y. L. Huang, Y. C. Chao, K. T. Tsai, W. C. Cheng, C. Y. Chuang, Y. H. Tsai, C. Y. Huang, D. W. Wang, C. H. Lin, J. K. Wang and Y. L. Wang, Differentiation of Bacteria Cell Wall Using Raman Scattering Enhanced by Nanoparticle Array, J. Nanosci. Nanotechnol., 2012, 12, 5004–5008 CrossRef CAS PubMed.
- C. Mariette, E. Tavernier, D. Hocquet, A. Huynh, F. Isnard, F. Legrand, V. Lhéritier, E. Raffoux, H. Dombret, N. Ifrah, J.-Y. Cahn and A. Thiébaut, Epidemiology of invasive fungal infections during induction therapy in adults with acute lymphoblastic leukemia: a GRAALL-2005 study, Leuk. Lymphoma, 2016, 1–8 Search PubMed.
- J. Timsit, E. Azoulay and C. Schwebel, et al., Empirical micafungin treatment and survival without invasive fungal infection in adults with icu-acquired sepsis, candida colonization, and multiple organ failure: The empiricus randomized clinical trial, J. Am. Med. Assoc., 2016, 316, 1555–1564 CrossRef CAS PubMed.
- J. Popp and M. Bauer, in Modern Techniques for Pathogen Detection, Wiley Vch, Weinheim, Germany, 2015, ch. 6, pp. 253–288 Search PubMed.
- V. Garzón, R.-H. Bustos and D. G. Pinacho, Personalized Medicine for Antibiotics: The Role of Nanobiosensors in Therapeutic Drug Monitoring, J. Pers. Med., 2020, 10, 147 CrossRef PubMed.
- J. Deng, S. Zhao, Y. Liu, C. Liu and J. Sun, Nanosensors for Diagnosis of Infectious Diseases, ACS Appl. Bio Mater., 2021, 4, 3863–3879 CrossRef CAS.
- S. Xu, J. Wang, Y. Zou, H. Liu, G. Wang, X. Zhang, S. Jiang, Z. Li, D. Cao and R. Tang, High performance SERS active substrates fabricated by directly growing graphene on Ag nanoparticles, RSC Adv., 2015, 5, 90457–90465 RSC.
- X. Meng, H. Wang, N. Chen, P. Ding, H. Shi, X. Zhai, Y. Su and Y. He, A Graphene-Silver Nanoparticle-Silicon Sandwich SERS Chip for Quantitative Detection of Molecules and Capture, Discrimination, and Inactivation of Bacteria, Anal. Chem., 2018, 90, 5646–5653 CrossRef CAS PubMed.
- X. Li, J. Li, X. Zhou, Y. Ma, Z. Zheng, X. Duan and Y. Qu, Silver nanoparticles protected by monolayer graphene as a stabilized substrate for surface enhanced Raman spectroscopy, Carbon, 2014, 66, 713–719 CrossRef CAS.
- W. Song, D. W. Li, Y. T. Li, Y. Li and Y. T. Long, Disposable biosensor based on graphene oxide conjugated with tyrosinase assembled gold nanoparticles, Biosens. Bioelectron., 2011, 26, 3181–3186 CrossRef CAS PubMed.
- Y. Li, J. Yang, T. Zhong, N. Zhao, Q.-Q. Liu, H.-F. Shi and H.-M. Xu, Fast and green synthesis of silver nanoparticles/reduced graphene oxide composite as efficient surface-enhanced Raman scattering substrate for bacteria detection, Monatsh. Chem., 2017, 148, 1155–1163 CrossRef CAS.
- X. Ling, L. Xie, Y. Fang, H. Xu, H. Zhang, J. Kong, M. S. Dresselhaus, J. Zhang and Z. Liu, Can Graphene be used as a Substrate for Raman Enhancement?, Nano Lett., 2010, 10, 553–561 CrossRef CAS PubMed.
- P. V. Kamat, Graphene-Based Nanoarchitectures. Anchoring Semiconductor and Metal Nanoparticles on a Two-Dimensional Carbon Support, J. Phys. Chem. Lett., 2010, 1, 520–527 CrossRef CAS.
- S. K. Srivastava, H. B. Hamo, A. Kushmaro, R. S. Marks, C. Gruner, B. Rauschenbach and I. Abdulhalim, Highly sensitive and specific detection of E. coli by a SERS nanobiosensor chip utilizing metallic nanosculptured thin films, Analyst, 2015, 140, 3201–3209 RSC.
- H. Etayash, K. Jiang, T. Thundat and K. Kaur, Impedimetric Detection of Pathogenic Gram-positive Bacteria Using an Antimicrobial Peptide from Class IIa Bacteriocins, Anal. Chem., 2014, 86, 1693–1700 CrossRef CAS PubMed.
- M. Hoyos-Nogués, S. Brosel-Oliu, N. Abramova, F.-X. Muñoz, A. Bratov, C. Mas-Moruno and F.-J. Gil, Impedimetric antimicrobial peptide-based sensor for the early detection of periodontopathogenic bacteria, Biosens. Bioelectron., 2016, 86, 377–385 CrossRef PubMed.
- X. Zhang, H. Zhang, S. Yan, Z. Zeng, A. Huang, A. Liu, Y. Yuan and Y. Huang, Organic Molecule Detection Based on SERS in Microfluidics, Sci. Rep., 2019, 9, 17634 CrossRef PubMed.
- W. Wang, V. Hynninen, L. Qiu, A. Zhang, T. Lemma, N. Zhang, H. Ge, J. J. Toppari, V. P. Hytönen and J. Wang, Synergistic enhancement via plasmonic nanoplate-bacteria-nanorod supercrystals for highly efficient SERS sensing of food-borne bacteria, Sens. Actuators, B, 2017, 239, 515–525 CrossRef CAS.
- Q. Zhou and T. Kim, Review of microfluidic approaches for surface-enhanced Raman scattering, Sens. Actuators, B, 2016, 227, 504–514 CrossRef CAS.
- H. Etayash, L. Norman, T. Thundat, M. Stiles and K. Kaur, Surface-Conjugated Antimicrobial Peptide Leucocin A Displays High Binding to Pathogenic Gram-positive Bacteria, ACS Appl. Mater. Interfaces, 2014, 6, 1131–1138 CrossRef CAS PubMed.
- Y. Shen, J. Yue, W. Xu and S. Xu, Recent progress of surface-enhanced Raman spectroscopy for subcellular compartment analysis, Theranostics, 2021, 11, 4872–4893 CrossRef PubMed.
- P. Wang, Y. Sun, X. Li, L. Wang, Y. Xu, L. He and G. Li, Recent advances in dual recognition based surface enhanced Raman scattering for pathogenic bacteria detection: A review, Anal. Chim. Acta, 2021, 1157, 338279 CrossRef CAS PubMed.
- K. Eberhardt, C. Stiebing, C. Matthäus, M. Schmitt and J. Popp, Advantages and limitations of Raman spectroscopy for molecular diagnostics: an update, Expert Rev. Mol. Diagn., 2015, 15, 773–787 CrossRef CAS PubMed.
- S. Pahlow, S. Meisel, D. Cialla-May, K. Weber, P. Rösch and J. Popp, Isolation and identification of bacteria by means of Raman spectroscopy, Adv. Drug Delivery Rev., 2015, 89, 105–120 CrossRef CAS PubMed.
- V. Kumar, B. Kampe, P. Rösch and J. Popp, Classification and identification of pigmented cocci bacteria relevant to the soil environment via Raman spectroscopy, Environ. Sci. Pollut. Res., 2015, 22, 19317–19325 CrossRef CAS PubMed.
- S. Hanf, T. Bögözi, R. Keiner, T. Frosch and J. Popp, Fast and Highly Sensitive Fiber-Enhanced Raman Spectroscopic Monitoring of Molecular H2 and CH4 for Point-of-Care Diagnosis of Malabsorption Disorders in Exhaled Human Breath, Anal. Chem., 2015, 87, 982–988 CrossRef CAS PubMed.
- P. Heraud, B. R. Wood, J. Beardall and D. McNaughton, Effects of pre-processing of Raman spectra on in vivo classification of nutrient status of microalgal cells, J. Chemom., 2006, 20, 193–197 CrossRef CAS.
- P. Geladi, B. Sethson, J. Nyström, T. Lillhonga, T. Lestander and J. Burger, Chemometrics in spectroscopy: Part 2. Examples, Spectrochim. Acta, Part B, 2004, 59, 1347–1357 Search PubMed.
- M. Kemmler, E. Rodner, P. Rösch, J. Popp and J. Denzler, Automatic identification of novel bacteria using Raman spectroscopy and Gaussian processes, Anal. Chim. Acta, 2013, 794, 29–37 CrossRef CAS PubMed.
- A. Silge, W. Schumacher, P. Rösch, P. A. Da Costa Filho, C. Gérard and J. Popp, Identification of water-conditioned Pseudomonas aeruginosa by Raman microspectroscopy on a single cell level, Syst. Appl. Microbiol., 2014, 37, 360–367 CrossRef CAS PubMed.
- A. A. Moawad, A. Silge, T. Bocklitz, K. Fischer, P. Rosch, U. Roesler, M. C. Elschner, J. Popp and H. Neubauer, A Machine Learning-Based Raman Spectroscopic Assay for the Identification of Burkholderia mallei and Related Species, Molecules, 2019, 24, 4516 CrossRef CAS PubMed.
- S. Guo, P. Rösch, J. Popp and T. Bocklitz, Modified PCA and PLS: Towards a better classification in Raman spectroscopy–based biological applications, J. Chemom., 2019, 34, SI e3202 Search PubMed.
- C. N. Kotanen, L. Martinez, R. Alvarez and J. W. Simecek, Surface enhanced Raman scattering spectroscopy for detection and identification of microbial pathogens isolated from human serum, Sens. Biosens. Res., 2016, 8, 20–26 Search PubMed.
- T. R. Cundari, J. Deng, H. F. Pop and C. Sârbu, Structural Analysis of Transition Metal β-X Substituent Interactions. Toward the Use of Soft Computing Methods for Catalyst Modeling, J. Chem. Inf. Comput. Sci., 2000, 40, 1052–1061 CrossRef CAS PubMed.
- H. F. Pop, T. L. Pop and C. Sarbu, Assessment of Heart Disease using Fuzzy Classification Techniques, Sci. World J., 2001, 1, 369–390 CrossRef CAS PubMed.
- C. Sârbu and H. F. Pop, Fuzzy robust estimation of central location, Talanta, 2001, 54, 125–130 CrossRef.
- C. Sârbu, K. Zehl and J. W. Einax, Fuzzy divisive hierarchical clustering of soil data using Gustafson–Kessel algorithm, Chemom. Intell. Lab. Syst., 2007, 86, 121–129 CrossRef.
- C. Sârbu and J. W. Einax, Study of traffic-emitted lead pollution of soil and plants using different fuzzy clustering algorithms, Anal. Bioanal. Chem., 2008, 390, 1293–1301 CrossRef PubMed.
- S. Butaciu, M. Senila, C. Sarbu, M. Ponta, C. Tanaselia, O. Cadar, M. Roman, E. Radu, M. Sima and T. Frentiu, Chemical modeling of groundwater in the Banat Plain, southwestern Romania, with elevated As content and co-occurring species by combining diagrams and unsupervised multivariate statistical approaches, Chemosphere, 2017, 172, 127–137 CrossRef CAS PubMed.
- H. F. Pop, J. W. Einax and C. Sârbu, Classical and fuzzy principal component analysis of some environmental samples concerning the pollution with heavy metals, Chemom. Intell. Lab. Syst., 2009, 97, 25–32 CrossRef CAS.
- N. E. Dina, A. M. R. Gherman, A. Colnişă, D. Marconi and C. Sârbu, Fuzzy characterization and classification of bacteria species detected at single-cell level by surface-enhanced Raman scattering, Spectrochim. Acta, Part A, 2021, 247, 119149 CrossRef CAS PubMed.
- S. Goyal, S. Kumar, M. A. Zaveri and A. K. Shukla, Fuzzy Similarity Measure Based Spectral Clustering Framework for Noisy Image Segmentation, Int. J. Uncertainty Fuzziness Knowl.-Based Syst, 2017, 25, 649–673 CrossRef.
- P. Bo, S. Fenzhen and M. Yunshan, A Cloud and Cloud Shadow Detection Method Based on Fuzzy c-Means Algorithm, IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens., 2020, 13, 1714–1727 Search PubMed.
- B. A. Beirami and M. Mokhtarzade, An automatic method for unsupervised feature selection of hyperspectral images based on fuzzy clustering of bands, Trait. Signal, 2020, 37, 319–324 CrossRef.
- D. Radziuk and H. Moehwald, Prospects for plasmonic hot spots in single molecule SERS towards the chemical imaging of live cells, Phys. Chem. Chem. Phys., 2015, 17, 21072–21093 RSC.
- M. S. Hizir, N. M. Robertson, M. Balcioglu, E. Alp, M. Rana and M. V. Yigit, Universal sensor array for highly selective system identification using two-dimensional nanoparticles, Chem. Sci., 2017, 8, 5735–5745 RSC.
- Z. Chen, T. Dodig-Crnković, J. M. Schwenk and S.-C. Tao, Current applications of antibody microarrays, Clin. Proteomics, 2018, 15, 7–7 CrossRef PubMed.
- S. Berus, E. Witkowska, K. Niciński, E. Sadowy, W. Puzia, P. Ronkiewicz and A. Kamińska, Surface-enhanced Raman scattering as a discrimination method of Streptococcus spp. and alternative approach for identifying capsular types of S. pneumoniae isolates, Spectrochim. Acta, Part A, 2020, 118088, DOI:10.1016/j.saa.2020.118088.
- K. Kneipp, H. Kneipp, V. B. Kartha, R. Manoharan, G. Deinum, I. Itzkan, R. R. Dasari and M. S. Feld, Detection and identification of a single DNA base molecule using surface-enhanced Raman scattering (SERS), Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top., 1998, 57, R6281–R6284 CrossRef CAS.
- M. M. Harper, J. A. Dougan, N. C. Shand, D. Graham and K. Faulds, Detection of SERS active labelled DNA based on surface affinity to silver nanoparticles, Analyst, 2012, 137, 2063–2068 RSC.
- K. Gracie, V. Dhamodharan, P. I. Pradeepkumar, K. Faulds and D. Graham, Qualitative SERS analysis of G-quadruplex DNAs using selective stabilising ligands, Analyst, 2014, 139, 4458–4465 RSC.
- A. Torres-Nuñez, K. Faulds, D. Graham, R. A. Alvarez-Puebla and L. Guerrini, Silver colloids as plasmonic substrates for direct label-free surface-enhanced Raman scattering analysis of DNA, Analyst, 2016, 141, 5170–5180 RSC.
- R. Pilot, R. Signorini, C. Durante, L. Orian, M. Bhamidipati and L. Fabris, A Review on Surface-Enhanced Raman Scattering, Biosensors, 2019, 9, 57 CrossRef CAS PubMed.
- G. Barzan, A. Sacco, L. Mandrile, A. M. Giovannozzi, J. Brown, C. Portesi, M. R. Alexander, P. Williams, K. R. Hardie and A. M. Rossi, New frontiers against antibiotic resistance: A Raman-based approach for rapid detection of bacterial susceptibility and biocide-induced antibiotic cross-tolerance, Sens. Actuators, B, 2020, 309, 127774 CrossRef CAS.
- U.-C. Schröder, A. Ramoji, U. Glaser, S. Sachse, C. Leiterer, A. Csaki, U. Hübner, W. Fritzsche, W. Pfister, M. Bauer, J. Popp and U. Neugebauer, Combined Dielectrophoresis–Raman Setup for the Classification of Pathogens Recovered from the Urinary Tract, Anal. Chem., 2013, 85, 10717–10724 CrossRef PubMed.
- J. Kirchhoff, U. Glaser, J. A. Bohnert, M. W. Pletz, J. Popp and U. Neugebauer, Simple Ciprofloxacin Resistance Test and Determination of Minimal Inhibitory Concentration within 2 h Using Raman Spectroscopy, Anal. Chem., 2018, 90, 1811–1818 CrossRef CAS PubMed.
- A. Colniţă, N. E. Dina, N. Leopold, D. C. Vodnar, D. Bogdan, S. A. Porav and L. David, Characterization and Discrimination of Gram-positive Bacteria Using Raman Spectroscopy with the Aid of Principal Component Analysis, Nanomaterials, 2017, 7, 248 CrossRef PubMed.
- S. Fu, X. Wang, T. Wang, Z. Li, D. Han, C. Yu, C. Yang, H. Qu, H. Chi, Y. Wang, S. Li, B. Tian, W. Li and Z. Xia, A sensitive and rapid bacterial antibiotic susceptibility test method by surface enhanced Raman spectroscopy, Braz. J. Microbiol., 2020, 51, 875–881 CrossRef CAS PubMed.
- D. Bauer, K. Wieland, L. Qiu, A. C. Neumann-Cip, G. Magistro, C. Stief, A. Wieser and C. Haisch, Heteroresistant Bacteria Detected by an Extended Raman-Based Antibiotic Susceptibility Test, Anal. Chem., 2020, 92, 8722–8731 CrossRef CAS PubMed.
- A. Gupta, L. Holoidovsky, C. Thamaraiselvan, A. K. Thakur, S. P. Singh, M. M. Meijler and C. J. Arnusch, Silver-doped laser-induced graphene for potent surface antibacterial activity and anti-biofilm action, Chem. Commun., 2019, 55, 6890–6893 RSC.
- A. M. R. Gherman, N. E. Dina, V. Chiş, A. Wieser and C. Haisch, Yeast cell wall – Silver nanoparticles interaction: A synergistic approach between surface-enhanced Raman scattering and computational spectroscopy tools, Spectrochim. Acta, Part A, 2019, 222, 117223 CrossRef CAS PubMed.
- J. Xu, J. W. Turner, M. Idso, S. V. Biryukov, L. Rognstad, H. Gong, V. L. Trainer, M. L. Wells, M. S. Strom and Q. Yu, In Situ Strain-Level Detection and Identification of Vibrio parahaemolyticus Using Surface-Enhanced Raman Spectroscopy, Anal. Chem., 2013, 85, 2630–2637 CrossRef CAS PubMed.
- J. Xu, J. W. Turner, M. Idso, S. V. Biryukov, L. Rognstad, H. Gong, V. L. Trainer, M. L. Wells, M. S. Strom and Q. Yu, In situ strain-level detection and identification of Vibrio parahaemolyticus using surface-enhanced Raman spectroscopy, Anal. Chem., 2013, 85, 2630–2637 CrossRef CAS PubMed.
- N. P. Ivleva, M. Wagner, A. Szkola, H. Horn, R. Niessner and C. Haisch, Label-Free in Situ SERS Imaging of Biofilms, J. Phys. Chem. B, 2010, 114, 10184–10194 CrossRef CAS PubMed.
- A. Walter, A. März, W. Schumacher, P. Rösch and J. Popp, Towards a fast, high specific and reliable discrimination of bacteria on strain level by means of SERS in a microfluidic device, Lab Chip, 2011, 11, 1013–1021 RSC.
- T.-Y. Liu, K.-T. Tsai, H.-H. Wang, Y. Chen, Y.-H. Chen, Y.-C. Chao, H.-H. Chang, C.-H. Lin, J.-K. Wang and Y.-L. Wang, Functionalized arrays of Raman-enhancing nanoparticles for capture and culture-free analysis of bacteria in human blood, Nat. Commun., 2011, 2, 538 CrossRef PubMed.
- Y. Wang and S. Schlücker, Rational design and synthesis of SERS labels, Analyst, 2013, 138, 2224–2238 RSC.
- X. Xu, X. Ma, H. Wang and Z. Wang, Aptamer based SERS detection of Salmonella typhimurium using DNA-assembled gold nanodimers, Microchim. Acta, 2018, 185, 325 CrossRef PubMed.
- X. Zhang, S. Liu, X. Song, H. Wang, J. Wang, Y. Wang, J. Huang and J. Yu, Robust and universal SERS sensing platform for multiplexed detection of Alzheimer's disease core biomarkers using PAapt-AuNPs conjugates, ACS Sens., 2019, 4, 2140–2149 CrossRef CAS PubMed.
- G. Azemtsop Matanfack, A. Pistiki, P. Rösch and J. Popp, Raman 18O-Labeling of Bacteria in Visible and Deep UV-Ranges, J. Biophotonics, 2021, e202100013 Search PubMed.
- W. Zhu, C.-Y. Wang, J.-M. Hu and A.-G. Shen, Promoted “Click” SERS Detection for Precise Intracellular Imaging of Caspase-3, Anal. Chem., 2021, 93, 4876–4883 CrossRef CAS PubMed.
- M. Potara, A. Campu, D. Maniu, M. Focsan, I. Botiz and S. Astilean, in Advanced Nanostructures for Environmental Health, ed. L. Baia, Z. Pap, K. Hernadi and M. Baia, Elsevier, 2020, pp. 347–384 Search PubMed.
- W. J. Thrift, S. Ronaghi, M. Samad, H. Wei, D. G. Nguyen, A. S. Cabuslay, C. E. Groome, P. J. Santiago, P. Baldi and A. I. Hochbaum, Deep Learning Analysis of Vibrational Spectra of Bacterial Lysate for Rapid Antimicrobial Susceptibility Testing, ACS Nano, 2020, 14, 15336–15348 CrossRef PubMed.
- X. Qiao, B. Su, C. Liu, Q. Song, D. Luo, G. Mo and T. Wang, Selective Surface Enhanced Raman Scattering for Quantitative Detection of Lung Cancer Biomarkers in Superparticle@MOF Structure, Adv. Mater., 2018, 30, 1702275 CrossRef PubMed.
- X. Fang, Q. Zeng, X. Yan, Z. Zhao, N. Chen, Q. Deng, M. Zhu, Y. Zhang and S. Li, Fast discrimination of tumor and blood cells by label-free surface-enhanced Raman scattering spectra and deep learning, J. Appl. Phys., 2021, 129, 123103 CrossRef CAS.
- Z. Fan, R. Kanchanapally and P. C. Ray, Hybrid Graphene Oxide Based Ultrasensitive SERS Probe for Label-Free Biosensing, J. Phys. Chem. Lett., 2013, 4, 3813–3818 CrossRef CAS.
- I. Bruzas, W. Lum, Z. Gorunmez and L. Sagle, Advances in surface-enhanced Raman spectroscopy (SERS) substrates for lipid and protein characterization: sensing and beyond, Analyst, 2018, 143, 3990–4008 RSC.
- H. J. Park, S. Cho, M. Kim and Y. S. Jung, Carboxylic Acid-Functionalized, Graphitic Layer-Coated Three-Dimensional SERS Substrate for Label-Free Analysis of Alzheimer's Disease Biomarkers, Nano Lett., 2020, 20, 2576–2584 CrossRef CAS PubMed.
- D. Sun, F. Cao, Y. Tian, A. Li, W. Xu, Q. Chen, W. Shi and S. Xu, Label-free detection of multiplexed metabolites at single-cell level via a SERS-microfluidic droplet platform, Anal. Chem., 2019, 91, 15484–15490 CrossRef CAS PubMed.
- S. Hu, F. Gu, M. Chen, C. Wang, J. Li, J. Yang, G. Wang, Z. Zhou and Y. Yang, A novel method for identifying and distinguishing Cryptococcus neoformans and Cryptococcus gattii by surface-enhanced Raman scattering using positively charged silver nanoparticles, Sci. Rep., 2020, 10, 12480 CrossRef CAS PubMed.
- G. Bodelón, V. Montes-García, J. Pérez-Juste and I. Pastoriza-Santos, Surface-Enhanced Raman Scattering Spectroscopy for Label-Free Analysis of P. aeruginosa Quorum Sensing, Front. Cell. Infect. Microbiol., 2018, 8 Search PubMed.
- Q. Zhu, H. Li, F. Lu, Y. Chai and Y. Yuan, A Widely Applicable Silver Sol for TLC Detection with Rich and Stable SERS Features, Nanoscale Res. Lett., 2016, 11, 220 CrossRef PubMed.
- J. Lee, J. Park, A. Go, H. Moon, S. Kim, S. Jung, W. Jeong and H. Chung, Urine Multi-drug Screening with GC-MS or LC–MS-MS Using SALLE-hybrid PPT/SPE, J. Anal. Toxicol., 2018, 42, 617–624 CrossRef CAS PubMed.
- Y. Abiedalla, J. DeRuiter, F. Smith and C. R. Clark, Differentiation of the six dimethoxypyrovalerone regioisomers: GC-MS, GC-MS/MS and GC-IR, Talanta, 2017, 171, 220–228 CrossRef CAS PubMed.
- W. Li, Y. Wu, M. Wan, Y. Chu, X. Wang, S. Li, Z. Liu, X. Chen, N. Polachi and S. Zhou, Simultaneous determination of three saponins in human plasma after oral administration of compound danshen dripping pills by LC-MS/MS and its application in a pharmacokinetic study, J. Pharm. Biomed. Anal., 2019, 169, 254–259 CrossRef CAS PubMed.
- K. Guo, T. Zhang, Y. Wang, B. Jin and C. Ma, Characterization of degradation products and process-related impurity of sutezolid by liquid chromatography/electrospray ionization tandem mass spectrometry, J. Pharm. Biomed. Anal., 2019, 169, 196–207 CrossRef CAS PubMed.
- U. B. Hezel and C. Zeiss, in Journal of Chromatography Library, ed. A. Zlatkis and R. E. Kaiser, Elsevier, 1977, vol. 9, pp. 147–188 Search PubMed.
- F. Pozzi, N. Shibayama, M. Leona and J. R. Lombardi, TLC–SERS study of Syrian rue (Peganum harmala) and its main alkaloid constituents, J. Raman Spectrosc., 2013, 44, 102–107 CrossRef CAS.
- P. A. Mosier-Boss, K. C. Sorensen, R. D. George, P. C. Sims and A. Obraztsova, Surface enhanced Raman scattering of bacteria using capped and uncapped silver nanoparticles, Spectrochim. Acta, Part A, 2020, 242, 118742 CrossRef CAS PubMed.
- H. Jin, J. Wang, S. Jin, L. Jiang and Y. Zou, Raman Spectroscopy of Potential Bio-hazards commonly found in Bio-aerosols, Spectrochim. Acta, Part A, 2020, 118753 CrossRef CAS PubMed.
- M. Mehta, Y. Liu, M. Waterland and G. Holmes, Characterization of the Degradation of Sheepskin by Monitoring Cytochrome c of Bacteria by Raman Spectroscopy, Anal. Lett., 2020, 1–18 Search PubMed.
- C. Wei, M. Li and X. Zhao, Surface-Enhanced Raman Scattering (SERS) With Silver Nano Substrates Synthesized by Microwave for Rapid Detection of Foodborne Pathogens, Front. Microbiol., 2018, 9, 2857 CrossRef PubMed.
- E. Witkowska, D. Korsak, A. Kowalska, M. Księżopolska-Gocalska, J. Niedziółka-Jönsson, E. Roźniecka, W. Michałowicz, P. Albrycht, M. Podrażka, R. Hołyst, J. Waluk and A. Kamińska, Surface-enhanced Raman spectroscopy introduced into the International Standard Organization (ISO) regulations as an alternative method for detection and identification of pathogens in the food industry, Anal. Bioanal. Chem., 2017, 409, 1555–1567 CrossRef CAS PubMed.
- Q. Ding, Y. Kang, W. Li, G. Sun, H. Liu, M. Li, Z. Ye, M. Zhou, J. Zhou and S. Yang, Bioinspired Brochosomes as Broadband and Omnidirectional Surface-Enhanced Raman Scattering Substrates, J. Phys. Chem. Lett., 2019, 10, 6484–6491 CrossRef CAS PubMed.
- C. Catala, B. Mir-Simon, X. Feng, C. Cardozo, N. Pazos-Perez, E. Pazos, S. Gómez-de Pedro, L. Guerrini, A. Soriano, J. Vila, F. Marco, E. Garcia-Rico and R. A. Alvarez-Puebla, Online SERS Quantification of Staphylococcus aureus and the Application to Diagnostics in Human Fluids, Adv. Mater. Technol., 2016, 1, 1600163 CrossRef.
- L. Bi, X. Wang, X. Cao, L. Liu, C. Bai, Q. Zheng, J. Choo and L. Chen, SERS-active Au@Ag core-shell nanorod (Au@AgNR) tags for ultrasensitive bacteria detection and antibiotic-susceptibility testing, Talanta, 2020, 220, 121397 CrossRef CAS PubMed.
- H. Kearns, R. Goodacre, L. E. Jamieson, D. Graham and K. Faulds, SERS Detection of Multiple Antimicrobial-Resistant Pathogens Using Nanosensors, Anal. Chem., 2017, 89, 12666–12673 CrossRef CAS PubMed.
- S. Zhang, X. Tang, H. Zheng, D. Wang, Z. Xie, W. Ding and X. Zheng, Combination of bacitracin-based flocculant and surface enhanced Raman scattering labels for flocculation, identification and sterilization of multiple bacteria in water treatment, J. Hazard. Mater., 2020, 124389, DOI:10.1016/j.jhazmat.2020.124389.
- Q. Lv, H. Min, D.-B. Duan, W. Fang, G.-M. Pan, A.-G. Shen, Q.-Q. Wang, G. Nie and J.-M. Hu, Total Aqueous Synthesis of Au@Cu2−xS Core–Shell Nanoparticles for In Vitro and In Vivo SERS/PA Imaging-Guided Photothermal Cancer Therapy, Adv. Healthcare Mater., 2019, 8, 1801257 Search PubMed.
- E. Garai, S. Sensarn, C. L. Zavaleta, N. O. Loewke, S. Rogalla, M. J. Mandella, S. A. Felt, S. Friedland, J. T. C. Liu, S. S. Gambhir and C. H. Contag, A Real-Time Clinical Endoscopic System for Intraluminal, Multiplexed Imaging of Surface-Enhanced Raman Scattering Nanoparticles, PLoS One, 2015, 10, e0123185 CrossRef PubMed.
- Y. W. Wang, S. Kang, A. Khan, P. Q. Bao and J. T. C. Liu, In vivo multiplexed molecular imaging of esophageal cancer via spectral endoscopy of topically applied SERS nanoparticles, Biomed. Opt. Express, 2015, 6, 3714–3723 CrossRef CAS PubMed.
- S. Meisel, S. Stöckel, P. Rösch and J. Popp, Identification of meat-associated pathogens via Raman microspectroscopy, Food Microbiol., 2014, 38, 36–43 CrossRef CAS PubMed.
- U. Munchberg, P. Rosch, M. Bauer and J. Popp, Raman spectroscopic identification of single bacterial cells under antibiotic influence, Anal. Bioanal. Chem., 2014, 406, 3041–3050 CrossRef PubMed.
- C.-S. Ho, N. Jean, C. A. Hogan, L. Blackmon, S. S. Jeffrey, M. Holodniy, N. Banaei, A. A. E. Saleh, S. Ermon and J. Dionne, Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning, Nat. Commun., 2019, 10, 4927 CrossRef PubMed.
- S. Yerolatsitis, F. Yu, S. McAughtrie, M. G. Tanner, H. Fleming, J. M. Stone, C. J. Campbell, T. A. Birks and J. C. Knight, Ultra-low background Raman sensing using a negative-curvature fibre and no distal optics, J. Biophotonics, 2019, 12, e201800239 CrossRef PubMed.
- WHO, Scientific Brief- Advice on the use of point-of-care immunodiagnostic tests for COVID-19, https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19.
- A. N. Kuzmin, A. Pliss and P. N. Prasad, Ramanomics: New Omics Disciplines Using Micro Raman Spectrometry with Biomolecular Component Analysis for Molecular Profiling of Biological Structures, Biosensors, 2017, 7, 52 CrossRef PubMed.
- L. Zhang, M. J. Henson and S. S. Sekulic, Multivariate data analysis for Raman imaging of a model pharmaceutical tablet, Anal. Chim. Acta, 2005, 545, 262–278 CrossRef CAS.
Footnote |
| † These authors contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2021 |