Hyaluronic acid-based extracellular matrix triggers spontaneous M2-like polarity of monocyte/macrophage†
Hyebin
Kim
a,
Junghwa
Cha
ab,
Minjeong
Jang
a and
Pilnam
Kim
 *ab
*ab
aDepartment of Bio and Brain Engineering, Korea Advanced Institute of Science and Technology, Daejeon 34141, Republic of Korea. E-mail: pkim@kaist.ac.kr
bKAIST Institute for Health Science and Technology, Daejeon, Korea
First published on 5th March 2019
Abstract
Hyaluronic acid (HA) is found in various tumor tissues, and is considered tumor-associated extracellular matrix (ECM). Within this tumor-associated ECM, stromal cells, especially immune cells, are involved in tumor progression. However, the effects of tumor-associated ECM on the characteristics of immune cells remain unexplored. Therefore, we studied the triggering effect of HA on spontaneous M2-like polarity of monocytes/macrophages using HA-mixed collagen (HA-COL) matrix. In the presence of HA, expression of the HA receptor (CD44) and M2 polarity-related genes was upregulated in human monocytes (THP-1 cells). We confirmed the CD44-mediated activation of STAT3 in THP-1 cells cultured in an HA-rich environment. Furthermore, when we induced the THP-1 cells to differentiate into cells with M1 or M2 polarity within an HA-rich environment, the HA-rich environment influenced the direction of induction. Our findings might improve understanding of the crosstalk between immune cells and tumor-associated ECM, and facilitate development of tumor immunotherapy strategies.
In the tumor microenvironment, complex crosstalk occurs between the cellular compartment, which contains cancer and stromal cells, and the non-cellular compartment.1 A prominent non-cellular component, the extracellular matrix (ECM), is a complicated network of macromolecules with distinct properties.2 The ECM is remodeled dynamically by changing its composition, stiffness, and architecture.3,4 During tumor progression, the ECM is deregulated; collapse of ECM homeostasis results in an aberrant ECM environment. The remodeled ECM plays a pivotal role in providing biochemical and biophysical cues to surrounding cells, regulating tumor and stromal cells.2,4–6 Much recent evidence indicates that the interaction between the parenchyma ECM and stromal cells promotes tumor growth and progression.2,7–9
Hyaluronic acid (HA) is a long-chain polysaccharide that is a major component of the tumor-associated ECM, which is recognized as an important regulator of cancer progression.10–12 HA is overproduced and deposited in the ECM of the tumor microenvironment.10,13–15 The accumulation of HA leads to the recruitment of stromal cells, including monocytes, macrophages, fibroblasts, and endothelial cells, toward the tumor-associated ECM.16,17 HA accelerates the motility of stromal cells, causing them to infiltrate the tumor stroma.16,18 This modulates the behavior of the stromal cells, resulting in tumorigenesis, including invasion and angiogenesis.17
HA is an important immune regulator that affects the function of immune cells.19 HA activates immune cells and is involved in regulating tissue injury.20–22 In an HA-rich ECM microenvironment, monocytes/macrophages infiltrate the surrounding tumor stroma.16 Monocytes can have an immunosuppressive M2 polarity when exposed to soluble HA derived from a tumor.23,24 Furthermore, the expression of the HA receptor, CD44, is elevated in tumor-associated macrophages within the tumor-associated ECM.25 Indeed, the HA-associated microenvironment is involved in the formation of pro-tumor immunosuppressive macrophages.17,24 However, the effects of the HA-rich ECM on the behavior of immune cells are not clear.
To address this issue, we developed a three-dimensional (3D) HA matrix model of the tumor-associated ECM for inducing polarity of monocytes/macrophages. Using the HA-rich ECM-based 3D model, we verified the effects of the HA matrix on monocyte/macrophage polarity. We used the human monocyte cell line THP-1 and constructed a cell-laden HA-rich environment by embedding THP-1 cells within the matrix. In this approach, the HA-rich matrix is a non-soluble environmental factor that can be exploited to examine crosstalk between monocytes/macrophages and the tumor-associated ECM. We hypothesized that THP-1 cells cultured within an HA-mixed collagen (HA-COL) matrix spontaneously trigger M2-like polarity. To elucidate the effects of HA on polarity, we confirmed the expression of HA receptors and a polarity-related gene of monocytes/macrophages.
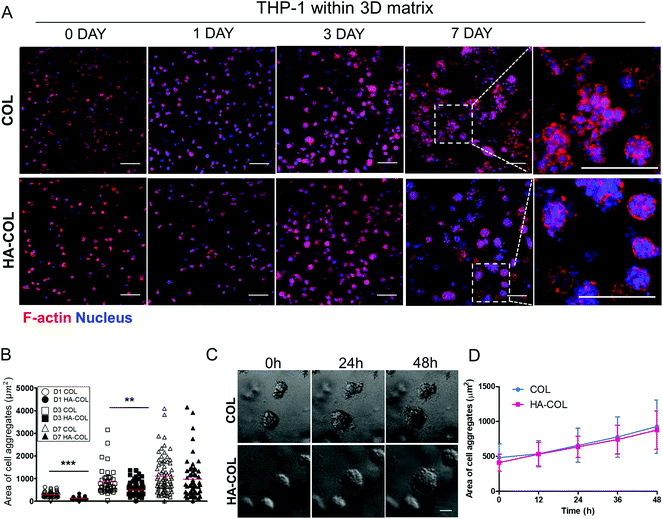
First, we cultured THP-1 cells in either collagen only (COL) or HA-COL matrix to observe their morphology and viability within the 3D matrix. After embedding dissociated THP-1 cells within both matrices, the F-actin and nuclei of the THP-1 cells were immunostained at 0, 1, 3, and 7 days to visualize the cell morphology. Over time, the THP-1 cells formed multi-cell aggregates within both 3D matrices (Fig. 1A). When we quantified the area of the THP-1 cell aggregates, the THP-1 cells cultured in the HA-COL matrix formed smaller aggregates than those in the COL matrix at 1 and 3 days (Fig. 1B). At 7 days, however, there was no significant difference in the size of the THP-1 cell aggregates between COL and HA-COL matrices. We examined the populations of live and dead THP-1 cells after culture in both 3D matrices for 7 days, and determined that the cell aggregates were mostly composed of live cells (Fig. S1†).
Next, we observed the formation of THP-1 cell aggregates using 48 h time-lapse imaging, to confirm whether the cell aggregates were proliferative. This showed that the cell aggregates in both the COL and HA-COL matrices grew due to proliferation, and not the clustering of smaller aggregates (Fig. 1C, ESI Videos 1 and 2†). Over 48 h, the size of the THP-1 aggregates in the HA-COL matrix increased from 410 to 878 μm2, while the size of the aggregates in the COL matrix increased from 482 to 928 μm2 (Fig. 1D). Unlike single cells or dispersed immune cells in two-dimensional (2D) cultures, we observed that a confined 3D environment enables both cell–cell and cell–ECM interactions, resulting in immune aggregates that have a morphology similar to that of human peripheral blood mononuclear cells.26
In addition, we confirmed the relative mRNA levels of HA synthesis (HAS1, HAS2) and HA degradation (HYAL1, HYAL2, HYAL3) by qRT-PCR (Fig. S2†). The metabolic-related gene expression showed no significant difference. It indicates that HA-COL could be hardly affected by the action of the cells. We further demonstrated the carbazole reaction based assay,27 which detects one of the components of the HA, glucuronic acid. By optical density (O.D.) analysis with culture media, we found that there was no difference in the concentration of solubilized HA between each of the groups according to cultivation time (Fig. S3B†). It indicates that the HA-COL nearly remain the stability during cultivation time with no remarkable degradation.
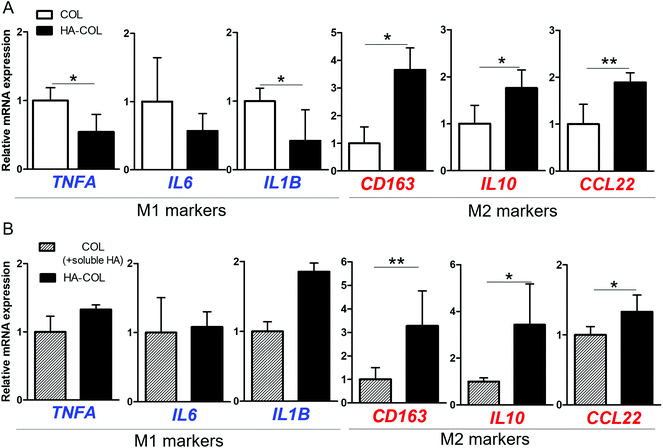
Next, we investigated the effect of the HA matrix on inducing polarity of THP-1 cells in the 3D microenvironment. THP-1 cells were cultured for 7 days in COL and HA-COL matrices, and changes in polarity were analyzed. As representative markers, we examine the expression of the pro-inflammatory cytokines tumor necrosis factor (TNF)-α, interleukin (IL)-6, and IL-1β to identify the M1 phenotype,28,29 and the immunosuppressive receptor, cytokine, and chemokines IL-10, CD163, and CCL22 for the M2 phenotype.28,30 For the M1 markers, the THP-1 cells grown in COL matrix expressed more TNFA, IL6, and IL1B than those in the HA-COL matrix (Fig. 2A). By contrast, regarding the M2 markers, the expression of CD163, IL10 and CCL22 was significantly upregulated in cells within the HA-COL matrix, by 3.7-, 1.8-, and 1.9-fold, respectively, compared with the cells within the COL matrix.
We then compared the polarity-related gene expression of THP-1 cells cultured within COL matrix supplemented with HA-containing culture medium (COL matrix + soluble HA) and HA-COL matrix, to confirm the effects of HA as a soluble factor regulating polarity. Regarding the M1 markers, there was no difference in the relative messenger RNA (mRNA) expression of the cells between the two matrices (Fig. 2B). By contrast, the expression of the M2 markers was significantly upregulated in the cells cultured in HA-COL matrix, compared with COL matrix containing soluble HA. The expression of the M2 markers CD163, IL10 and CCL22 of THP-1 cells in the HA-COL matrix was 3.3-, 3.4-, and 1.3-fold higher, respectively, than that of the cells within the COL matrix containing soluble HA. Therefore, the THP-1 cells grown in the HA-COL matrix showed elevated expression of M2 markers, indicating spontaneous induction of M2-like polarity in the HA-rich ECM environment. Moreover, the M2-like polarity of THP-1 cells was enhanced within the insoluble HA component incorporated into the crosslinked COL matrix compared with the COL matrix containing a solubilized HA component.
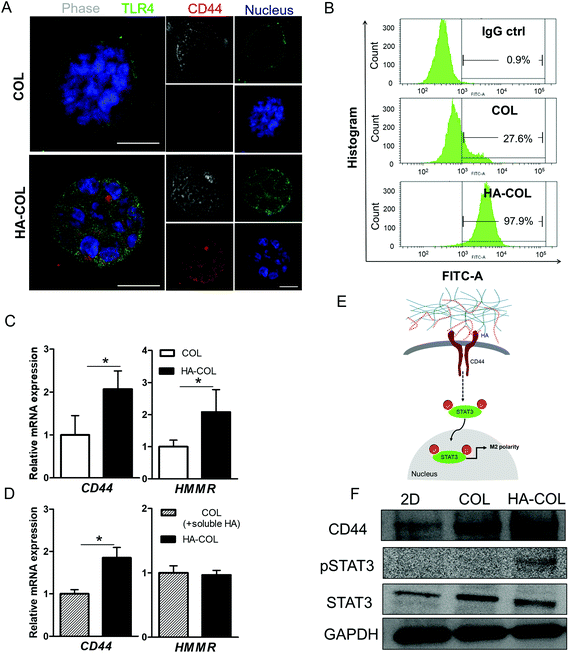
To investigate the mechanism underlying the spontaneous induction of M2-like polarity in the HA-COL matrix, we first confirmed the expression of the HA receptors CD44 and Toll-like receptor 4 (TLR4) to examine the receptor binding to the HA matrix.21,31 When we immunostained the THP-1 cells for CD44, TLR4, and nuclei, strong fluorescent signals from both CD44 and TLR4 were detected in cells in the HA-COL matrix compared with cells in the COL matrix (Fig. 3A). To quantify the CD44 expression of THP-1 cells, we analyzed the percentages of CD44-positive cells in the COL and HA-COL matrix by flow cytometry. As expected, the fraction of CD44-positive cells was significantly higher in the HA-COL matrix, by 3.5-fold, compared with the COL matrix (Fig. 3B). These results were confirmed in the gene expression analysis (Fig. 3C). The HA receptors CD44 and HA-mediated motility receptor (HMMR) were upregulated in THP-1 cells within HA-COL (both by 2.1-fold). Interestingly, depending on the HA solubility, the expression of CD44 and HMMR in THP-1 cells was 1.9-fold higher in the HA-COL matrix than in the COL matrix containing soluble HA (Fig. 3D). From these results, we confirmed that CD44 plays a prominent role as a major HA receptor in the HA-COL matrix.
Since CD44 is involved in regulating immune cell phenotypes, especially monocyte differentiation,32 we postulated that the elevated CD44 expression in the THP-1 cells within the HA-COL matrix was associated with the induction of M2 polarity via activation of the transcription factor signal transducer and activator of transcription-3 (STAT3). In fact, activation of STAT3 is strongly related to the regulation of macrophage polarity.33–35 Therefore, we verified the CD44-mediated activation of STAT3, as suggested in (Fig. 3E). Western blotting indicated that the phosphorylated form of STAT3 is elevated in the THP-1 cells grown in the HA-COL, along with increased expression of CD44 (Fig. 3F and Fig. S4†). This suggests that the induction of M2-like polarity in THP-1 cells is triggered by CD44-mediated STAT3 activation within an HA-rich ECM environment, directing cells toward an immunosuppressive, pro-tumorous phenotype.
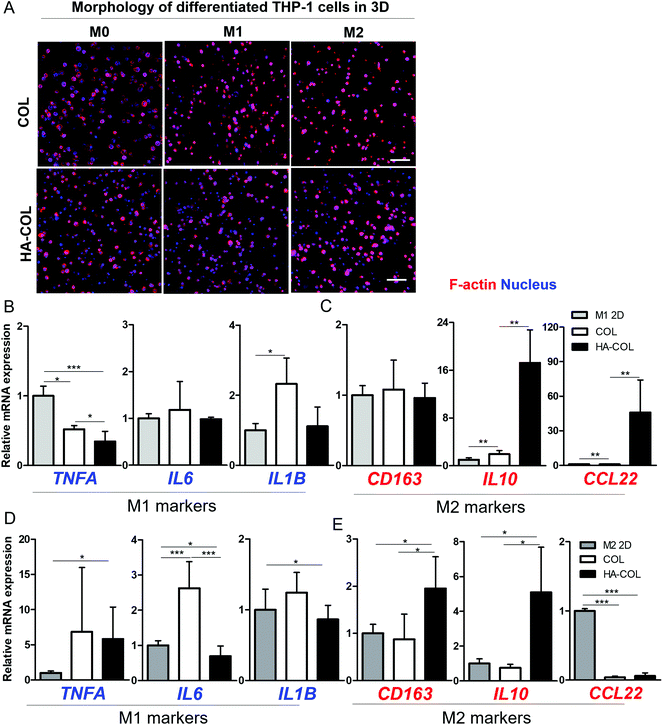
Finally, we confirmed the effect of the HA-rich ECM environment on regulating polarity under the chemical induction of THP-1 cells from monocytes to macrophages. For this purpose, we first treated the THP-1 cells with phorbol-12-myristate-13-acetate (PMA) (200 ng mL−1) for 48 h. Next, we incorporated lipopolysaccharide (LPS) (100 ng mL−1) for M1 polarization and IL-4/IL-13 (20 ng mL−1) for M2 polarization in culture medium respectively, following the conventional differentiation protocol.36,37 Compared with the morphology of THP-1 cells before polarization (M0) in a 2D environment, we observed that the morphologies of both M1- and M2-differentiated THP-1 cells in the 2D environment became elongated after full differentiation (Fig. S5†). In comparison, in both 3D matrices, both M1- and M2-differentiated THP-1 cells remained dissociated at the single-cell level after full differentiation (Fig. 4A), with high viability (Fig. S6†).
After the differentiation of THP-1 cells into M1 and M2 phenotypes, we investigated the polarity-related markers using gene expression analysis. For the M1-polarized THP-1 cells, the M1-polarity-related genes TNFA, IL6, and IL1B were downregulated within the HA-COL matrix compared with the COL matrix (Fig. 4B). In comparison, we observed upregulated expression of the M2-polarity-related genes CD163, IL10, and CCL22 in the HA-COL matrix (Fig. 4C). In M1-differentiated THP-1 cells, the relative mRNA expression levels of M1 markers (IL6 and IL1B) was downregulated 1.2- and 2.1-fold, and that of the M2 markers (IL10 and CCL22) was upregulated 8.8- and 48-fold in the HA-COL matrix. However, the M2-polairzed THP-1 cells expressed more M2 markers within the HA-COL matrix, but fewer M1 polarity markers, compared with THP-1 cells in COL matrix (Fig. 4D and E). With the differentiation of THP-1 cells into M2 polarity, the relative mRNA expression of M1 markers (IL6 and IL1B) was decreased 3.8- and 1.4-fold, and that of M2 markers (CD163, IL10, and CCL22) was upregulated 2.2-, 6.8-, and 1.6-fold, respectively, in the HA-COL matrix compared with the COL matrix. Consequently, we confirmed that even polarity-controlled THP-1 cells were strongly influenced by an HA-rich ECM environment, indicating that macrophage polarization to the M2 phenotype is dramatically enhanced in HA-COL despite induction of differentiation toward M1 polarity.
Based on our findings, an HA-rich ECM environment plays a crucial role in controlling macrophage polarity toward an immunosuppressive phenotype. Unlike most studies focusing on the role of tumor cells in regulating the immune polarity,23,38,39 our study addressed the importance of the ECM environment for tuning immune polarity. Therefore, our results are evidences for control of the polarity of immune cells via the interaction with the ECM environment, potentially suggesting new therapeutic targets for tumor-associated immune cells.
Conclusion
In this study, our ultimate objective was to verify the effects of an HA-rich ECM environment on regulating monocyte/macrophage polarity. Using a 3D HA-COL matrix, we demonstrated that THP-1 cells within a 3D HA-rich matrix environment show spontaneous M2-oriented polarity. Specifically, the main factor regulating the immune cell behavior, the highly upregulated CD44 of the THP-1 cells within the HA-rich ECM environment, triggered the activation of STAT3, inducing M2-like polarity of macrophages. In this regard, our results suggest that the HA environment induces a pro-tumor, immunosuppressive M2-like polarity in monocytes/macrophages. Our findings provide a better understanding of the crosstalk between an HA-rich ECM environment and immune cell phenotype. Since it is evident that HA influences the polarity of monocytes/macrophages,12 this suggests a new immunotherapeutic approach that involves altering the immune cell–ECM interaction.Experimental section
Cell culture
Human monocyte cell line, THP-1 cell line used in this study was acquired by Korean cell line bank. THP-1 cells were cultured in RPMI 1640 (Welgene, Korea) supplemented with 10% Fetal bovine serum (Welgene), 1% penicillin/streptomycin (Welgene). The cells were incubated at 37 °C in an atmosphere of 5% CO2 and 95% air. Cells were after 3 days and by centrifugation and splitting with 2.0 × 105 cell per ml to respective T 75 flask (SPL, Korea). For further experiments, cells were passaged and resuspended with full media and embedded within the 3D matrix by mixing with hydrogel solution or seeded in 2D cell culture plate with proper cell density.Preparation of COL and HA-COL hydrogel
We created the COL and HA-mixed collagen (HA-COL) hydrogel. For fabrication of COL hydrogel, high density collagen type I from rat tail (Corning) was diluted into 4.0 mg ml−1 collagen solutions in distilled water and 1 N NaOH was added to set the pH to 7.4 for cell culture. For HA-COL, the solution of sodium hyaluronate (Lifecore Biomedical, USA, 1.01–1.8 MDa) with concentration 4 mg ml−1 was used for diluting solvent, instead of distilled water used in the process of fabrication of COL hydrogel.40 Both hydrogels were incubated at 37 °C for 30 min and maintained at 37 °C in an atmosphere of 5% CO2 and 95% air following addition of the cell culture medium in hydrogel sample.Encapsulation of THP-1 cells within COL and HA-COL hydrogel with PDMS mold
To homogenous creation each of the sample, we used PDMS (Polydimethylsiloxane, Sylgard ® 184, Dow Corning) mold. PDMS was crossliked with curing agent by ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 and removed the bubbles with desiccator. After 1 h in oven, PDMS was punched with biopunch to fabricate the PDMS mold. Next, by using 3D matrices (COL and HA-COL hydrogel) which is explained above section, THP-1 cells were embedded in the 3D matrix. For elucidation effects of HA-rich tumorous environment on cell behavior, THP-1 cells were encapsulated in 3D matrix. THP-1 cells with appropriate density were passaged and prepared 750 cells per μl cell density to incorporate into 3D matrix. In COL hydrogel, THP-1 cells were resuspended with full media and mixed with COL hydrogel solution. Meanwhile, in HA-COL hydrogel, THP-1 cells were resuspended with 4 mg ml−1 HA-COL in replacement with full media and mixed with mixed with HA-COL hydrogel solution. With these solution, we seeded the hydrogel solution with THP-1 cells and did gelation in 37 °C for 30 min. Then, PDMS mold was took off and added culture medium in hydrogel sample. For analysis effects of HA solubility, we included the 100 μg ml−1 HA as soluble component in media. Then we incubated in 37 °C in an atmosphere of 5% CO2 and 95% air.
10 and removed the bubbles with desiccator. After 1 h in oven, PDMS was punched with biopunch to fabricate the PDMS mold. Next, by using 3D matrices (COL and HA-COL hydrogel) which is explained above section, THP-1 cells were embedded in the 3D matrix. For elucidation effects of HA-rich tumorous environment on cell behavior, THP-1 cells were encapsulated in 3D matrix. THP-1 cells with appropriate density were passaged and prepared 750 cells per μl cell density to incorporate into 3D matrix. In COL hydrogel, THP-1 cells were resuspended with full media and mixed with COL hydrogel solution. Meanwhile, in HA-COL hydrogel, THP-1 cells were resuspended with 4 mg ml−1 HA-COL in replacement with full media and mixed with mixed with HA-COL hydrogel solution. With these solution, we seeded the hydrogel solution with THP-1 cells and did gelation in 37 °C for 30 min. Then, PDMS mold was took off and added culture medium in hydrogel sample. For analysis effects of HA solubility, we included the 100 μg ml−1 HA as soluble component in media. Then we incubated in 37 °C in an atmosphere of 5% CO2 and 95% air.
Differentiation of THP-1 cells toward M1 and M2 polarity
THP-1 cells were seeded in dish or 24 well cell culture plate (SPL) then cells were treated with 200 ng ml−1 phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich) for 48 h then changed into full media and incubated during 24 h to 2D and 3D seeded sample respectively for resting. For M1 differentiation, cells were treated 100 ng ml−1 lipopolysaccharide (LPS) (Sigma-Aldrich) for 48 h. For M2 differentiation, cells were co treated 20 ng ml−1 interleukin (IL)-4 (Peprotech) and 20 ng ml−1 interleukin (IL)-13 (Peprotech) for at least 48 h.Immunocytochemistry (ICC)
Cells within 3D COL and HA-COL matrix for 7 days were fixed with 4% paraformaldehyde (Biosesang) in PBS for 15 min at room temperature. Then, cells were permeabilized with 0.3% Tirton X-100 (Sigma-Aldrich) for 15 min and were blocked with 1% Bovine serum albumin (BSA, Sigma-Aldrich) for 1 h at room temperature respectively. After treating each of the solutions, samples were washed 3 times with Dulbecco's Phosphate-Buffered Saline (DPBS) (Welgene). Cells were incubated with specific primary antibodies (anti-rabbit CD44 (Abcam), anti-mouse TLR4 (Santa cruz) overnight at 4 °C. After washing with DPBS at 3 times, cells were incubated with the secondary antibody for 3 h at room temperature. Then, cells were treated TRITC-phalloidin (Sigma-Aldrich) to stain F-actin and DAPI (Sigma-Aldrich) to stain nucleus respectively for 15 min at room temperature.Time-lapse imaging
For observation of morphology of THP-1 cells within both of the 3D matrices, we demonstrated the time-lapse imaging for 48 h by confocal microscope (NIKON, Japan). For maintain the cell culture condition, we used the Live chamber, which can keep in an atmosphere of 5% CO2 and 95% air at 37 °C. Then, we set the program for live imaging by NIS-Elements software.Quantitative real-time polymerase chain reaction (qRT-PCR)
For examination of the gene expression of THP-1 cells within COL and HA-COL matrix, cells were prepared for qRT-PCR after 7 days. For extraction of mRNA from the sample (at least n = 4–6 per each group) according to conventional RNA extraction protocol. By using TRIzol reagent (Ambion), cells were disrupted and mRNAs were extracted then cells were reacted chloroform (Sigma). After 15 min, mRNAs extracted from each sample were centrifugated at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min mRNAs were collected into new tubes for RNA precipitation by adding isopropanol (Sigma-Aldrich) and glycogen (Roche) within freezing state. Concentration of precipitated mRNAs was measured by microplate reader (Molecular device). Reverse transcription was carried out using the cDNA Synthesis kit (Bio-Rad) with PCR Master Mix (Bio-Rad). Then gene expression levels of each markers were quantified using signal amplification of SYBR green (Bio-rad) for markers listed on table. The gene expression levels were determined with the comparative Ct method in which the target genes were normalized to the endogenous reference (GAPDH). The forward/reverse sequences used for qRT-PCR were listed in Table 1.
000 rpm for 10 min mRNAs were collected into new tubes for RNA precipitation by adding isopropanol (Sigma-Aldrich) and glycogen (Roche) within freezing state. Concentration of precipitated mRNAs was measured by microplate reader (Molecular device). Reverse transcription was carried out using the cDNA Synthesis kit (Bio-Rad) with PCR Master Mix (Bio-Rad). Then gene expression levels of each markers were quantified using signal amplification of SYBR green (Bio-rad) for markers listed on table. The gene expression levels were determined with the comparative Ct method in which the target genes were normalized to the endogenous reference (GAPDH). The forward/reverse sequences used for qRT-PCR were listed in Table 1.
| Gene | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) |
|---|---|---|
| hGAPDH | GTATGACAACAGCCTCAAGAT | AGT CCT TCC ACG ATA CCA AA |
| hCD44 | CTCTCTCCCTCCACTTCAC | GCCTAATGTCCAGTTTCTTTCA |
| hRHAMM | GTTTCTGGAGCTGCTCCGTC | ACTGGTCCTTTCAATACTTCTAAAGT |
| hTNFα | ATGAGCACTGAAAGCATGATCC | GAGGGCTGATTAGAGAGAGGTC |
| hIL-1β | CAGCTACGAATCTCCGACCAC | GGCAGGGAACCAGCATCTTC |
| hIL-6 | AACCTGAACCTTCCAAAGATGG | CTGGCTTGTTCCTCACTACT |
| hCD163 | TTTGTCAACTTGAGTCCCTTCAC | TCCCGCTACACTTGTTTTCAC |
| hIL-10 | TCAAGGCGCATGTGAACTCC | GATGTCAAACTCACTCATGGCT |
| hCCL22 | ATCGCCTACAGACTGCACTC | GACGGTAACGGACGTAATCAC |
Flow cytometric (FACS) analysis
THP-1 cells in 2D were harvested by ultra centrifugation. THP-1 cells in 3D matrix were harvested by adding 200× collagenase (Thermo fisher) in medium for 1 h. Then cells, which is more than 1 × 106 cells per sample were washed FACS buffer (DPBS supplemented with 0.5% BSA and 0.1% sodium azide) for three times. Next, anti-rabbit CD44 (Abcam) and IgG isotype (Santa cruz) were treated to sample for 30 min at 4 °C. After ultra-centrifugation, samples were washed three times by resuspending the pellet with FACS buffer. Secondary antibody treated to samples for 30 min at 4 °C. Samples were washed three times with FACS buffer after ultracentrifugation by resuspension of cell pellet. By 4% paraformaldehyde (Biosesang), samples were fixed for 10 min at room temperature. Samples were analyzed by FACS (LSR Fortessa). Gates were determined by un-stained sample of THP-1 cells. Results were compared with samples with IgG isotypes control.Western blot analysis
Proteins were extracted from hydrogel with Protease inhibitor (100×, Thermo Scientific) and Phosphatase inhibitor (100×, Thermo Scientific) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 with RIPA buffer (Sigma-Aldrich) following ultra-centrifugation. For measurement of concentration of protein, Bradford assay was done by using 1
100 with RIPA buffer (Sigma-Aldrich) following ultra-centrifugation. For measurement of concentration of protein, Bradford assay was done by using 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 diluted protein assay dye reagent (Bio-rad). And then absorbance at 595 nm was measured with Microplate Reader (Molecular devices). Protein containing same amount of total protein were separated on 10% sodium dodecyl sulfate-poly acrylamide gel electrophoresis (SDS-PAGE) gels. Then separated proteins were transferred to the nitrocellulose membrane. The membrane was blocked with 5% Bovine serum albumin (BSA) (Sigma-Aldrich) and incubated with specific primary antibodies (CD44 (Abcam), GAPDH (Santa cruz) STAT3 (Santa cruz), pSTAT3 (Y705, Abcam)) at 4 °C. After washing 3 times with TBST solution, which contains Tris-buffered saline and TWEEN 20, membrane was incubated with horseradish peroxidase conjugated secondary antibodies for 1 h at room temperature. Membrane was washed three times with TBST solution and HRP-conjugated antibodies reacted with combined stable peroxide solution and luminol solution (Thermo Scientific) where reaction emitted light at 425 nm. CCD camera and phosphorimagers using chemiluminescence image analyzer (ImageQuant LAS 4000 mini, GE healthcare) captured emitted light. Quantification of band intensities was conducted using the ImageJ.
4 diluted protein assay dye reagent (Bio-rad). And then absorbance at 595 nm was measured with Microplate Reader (Molecular devices). Protein containing same amount of total protein were separated on 10% sodium dodecyl sulfate-poly acrylamide gel electrophoresis (SDS-PAGE) gels. Then separated proteins were transferred to the nitrocellulose membrane. The membrane was blocked with 5% Bovine serum albumin (BSA) (Sigma-Aldrich) and incubated with specific primary antibodies (CD44 (Abcam), GAPDH (Santa cruz) STAT3 (Santa cruz), pSTAT3 (Y705, Abcam)) at 4 °C. After washing 3 times with TBST solution, which contains Tris-buffered saline and TWEEN 20, membrane was incubated with horseradish peroxidase conjugated secondary antibodies for 1 h at room temperature. Membrane was washed three times with TBST solution and HRP-conjugated antibodies reacted with combined stable peroxide solution and luminol solution (Thermo Scientific) where reaction emitted light at 425 nm. CCD camera and phosphorimagers using chemiluminescence image analyzer (ImageQuant LAS 4000 mini, GE healthcare) captured emitted light. Quantification of band intensities was conducted using the ImageJ.
Statistical analysis
In all of the statistical analysis, normalization of values from independent experiments was presented as the mean ± standard error of the mean (SEM). Levels of significance for comparisons between independent samples were analyzed by using Student's t-test. Groups were compared by one-way analysis of variance (ANOVA) with Tukey's post hoc test applied to significant main effects. For denotation of statistically difference by p-values, asterisks (*) were considered when p-value is p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***). All of the analysis of gene expression level was confirmed through repetition at 3–5 times.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Prof. Suk-Jo Kang for discussions and insightful comments. This research was supported from the National Research Foundation of Korea (NRF) (grant number: NRF-2015H1A2A1030560) and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C1324).References
- X. Xu, M. C. Farach-Carson and X. Jia, Biotechnol. Adv., 2014, 32, 1256–1268 CrossRef CAS PubMed.
- P. Lu, V. M. Weaver and Z. Werb, J. Cell Biol., 2012, 196, 395–406 CrossRef CAS PubMed.
- T. R. Cox and J. T. Erler, Dis. Models Mech., 2011, 4, 165–178 CrossRef CAS PubMed.
- J. L. Leight, A. P. Drain and V. M. Weaver, Annu. Rev. Cancer Biol., 2017, 1, 313–334 CrossRef.
- M. Giussani, G. Merlino, V. Cappelletti, E. Tagliabue and M. G. Daidone, Semin. Cancer Biol., 2015, 35, 3–10 CrossRef CAS PubMed.
- M. W. Pickup, J. K. Mouw and V. M. Weaver, EMBO Rep., 2014, 15, 1243–1253 CrossRef CAS PubMed.
- A. W. Holle, J. L. Young and J. P. Spatz, Adv. Drug Delivery Rev., 2016, 97, 270–279 CrossRef CAS PubMed.
- K. Ghosh, C. K. Thodeti, A. C. Dudley, A. Mammoto, M. Klagsbrun and D. E. Ingber, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 11305–11310 CrossRef CAS PubMed.
- D. F. Quail and J. A. Joyce, Nat. Med., 2013, 19, 1423–1437 CrossRef CAS PubMed.
- T. Chanmee, P. Ontong and N. Itano, Cancer Lett., 2016, 375, 20–30 CrossRef CAS PubMed.
- B. P. Toole, Nat. Rev. Cancer, 2004, 4, 528–539 CrossRef CAS PubMed.
- A. Kultti, X. Li, P. Jiang, C. B. Thompson, G. I. Frost and H. M. Shepard, Cancers, 2012, 4, 873–903 CrossRef CAS PubMed.
- R. H. Tammi, A. Kultti, V. M. Kosma, R. Pirinen, P. Auvinen and M. I. Tammi, Semin. Cancer Biol., 2008, 18, 288–295 CrossRef CAS PubMed.
- N. Sato, S. Kohi, K. Hirata and M. Goggins, Cancer Sci., 2016, 107, 569–575 CrossRef CAS PubMed.
- X. B. Cheng, N. Sato, S. Kohi and K. Yamaguchi, PLoS One, 2013, 8, e80765 CrossRef CAS PubMed.
- N. Itano, L. Zhuo and K. Kimata, Cancer Sci., 2008, 99, 1720–1725 CrossRef CAS PubMed.
- N. Kobayashi, S. Miyoshi, T. Mikami, H. Koyama, M. Kitazawa, M. Takeoka, K. Sano, J. Amano, Z. Isogai, S. Niida, K. Oguri, M. Okayama, J. A. McDonald, K. Kimata, S. Taniguchi and N. Itano, Cancer Res., 2010, 70, 7073–7083 CrossRef CAS PubMed.
- E. A. Turley, P. W. Noble and L. Y. Bourguignon, J. Biol. Chem., 2002, 277, 4589–4592 CrossRef CAS PubMed.
- D. Nikitovic, M. Tzardi, A. Berdiaki, A. Tsatsakis and G. N. Tzanakakis, Front. Immunol., 2015, 6, 169 Search PubMed.
- C. Termeer, F. Benedix, J. Sleeman, C. Fieber, U. Voith, T. Ahrens, K. Miyake, M. Freudenberg, C. Galanos and J. C. Simon, J. Exp. Med., 2002, 195, 99–111 CrossRef CAS PubMed.
- D. Jiang, J. Liang, J. Fan, S. Yu, S. Chen, Y. Luo, G. D. Prestwich, M. M. Mascarenhas, H. G. Garg, D. A. Quinn, R. J. Homer, D. R. Goldstein, R. Bucala, P. J. Lee, R. Medzhitov and P. W. Noble, Nat. Med., 2005, 11, 1173–1179 CrossRef CAS PubMed.
- K. A. Scheibner, M. A. Lutz, S. Boodoo, M. J. Fenton, J. D. Powell and M. R. Horton, J. Immunol., 2006, 177, 1272–1281 CrossRef CAS.
- G. Zhang, L. Guo, C. Yang, Y. Liu, Y. He, Y. Du, W. Wang and F. Gao, OncoImmunology, 2016, 5, e1172154 CrossRef PubMed.
- D. M. Kuang, Y. Wu, N. Chen, J. Cheng, S. M. Zhuang and L. Zheng, Blood, 2007, 110, 587–595 CrossRef CAS PubMed.
- M. D. Duff, J. Mestre, S. Maddali, Z. P. Yan, P. Stapleton and J. M. Daly, J. Surg. Res., 2007, 142, 119–128 CrossRef CAS PubMed.
- E. M. Sturm, B. Radnai, K. Jandl, A. Stancic, G. P. Parzmair, C. Hogenauer, P. Kump, H. Wenzl, W. Petritsch, T. R. Pieber, R. Schuligoi, G. Marsche, N. Ferreiros, A. Heinemann and R. Schicho, J. Immunol., 2014, 193, 827–839 CrossRef CAS PubMed.
- M. Cesaretti, E. Luppi, F. Maccari and N. Volpi, Carbohydr. Polym., 2003, 54, 59–61 CrossRef CAS.
- N. Wang, H. Liang and K. Zen, Front. Immunol., 2014, 5, 614 Search PubMed.
- F. O. Martinez and S. Gordon, F1000Prime Rep., 2014, 6, 13 Search PubMed.
- D. M. Mosser and X. Zhang, Immunol. Rev., 2008, 226, 205–218 CrossRef CAS PubMed.
- H. Ponta, L. Sherman and P. A. Herrlich, Nat. Rev. Mol. Cell Biol., 2003, 4, 33–45 CrossRef CAS PubMed.
- G. Zhang, H. Zhang, Y. Liu, Y. He, W. Wang, Y. Du, C. Yang and F. Gao, Acta Biochim. Biophys. Sin., 2014, 46, 540–547 CrossRef CAS PubMed.
- T. Lawrence and G. Natoli, Nat. Rev. Immunol., 2011, 11, 750–761 CrossRef CAS PubMed.
- J. J. O'Shea, M. Pesu, D. C. Borie and P. S. Changelian, Nat. Rev. Drug Discovery, 2004, 3, 555–564 CrossRef PubMed.
- J. H. Lyu, B. Huang, D. W. Park and S. H. Baek, J. Cell. Biochem., 2016, 117, 483–490 CrossRef CAS PubMed.
- U. Saqib, S. Sarkar, K. Suk, O. Mohammad, M. S. Baig and R. Savai, Oncotarget, 2018, 9, 17937–17950 Search PubMed.
- J. G. Quatromoni and E. Eruslanov, Am. J. Transl. Res., 2012, 4, 376–389 Search PubMed.
- R. Noy and J. W. Pollard, Immunity, 2014, 41, 49–61 CrossRef CAS PubMed.
- H. He, S. Zhang, S. Tighe, J. Son and S. C. Tseng, J. Biol. Chem., 2013, 288, 25792–25803 CrossRef CAS PubMed.
- J. Cha, S. G. Kang and P. Kim, Sci. Rep., 2016, 6, 24912 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9bm00155g |
| This journal is © The Royal Society of Chemistry 2019 |