Editorial: Strategies for cellular target identification of natural products
D.
Romo
*a and
J. O.
Liu
*b
aBaylor University, USA. E-mail: Daniel_Romo@baylor.edu
bJohns Hopkins University, USA. E-mail: joliu@jhu.edu
The completion of the human genome project has led to the deciphering of the human genetic code and over 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 genes that define our physical existence. However, this fantastic advance only opened further questions and research directions, as the functions of the majority of human gene products remain unknown. In addition, to unravel the functions of those genes, a major task for chemical biologists will be to discover inhibitors and activators of these gene products, both as chemical probes and potential drug leads (chemical genetics).2 Recent studies have revealed that only 266 human genome-derived targets of the druggable genome (estimated to be ∼3000)3 are currently targeted by small molecules, with up to twice that number if one considers that proteins with 50% identity may display similar pharmacology. Thus, the dire need for discovery of small molecules that can modulate protein function to address human disease cannot be understated. Indeed, a reasonable extension of the central dogma of biology highlights the great importance of small molecules in biological information flow (Fig. 1) given their proven ability to modulate the activity of biological macromolecules (Fig. 1).4 Looking at this gap in knowledge regarding small molecule–protein interactions in a different way, the fact that only 130 ‘privileged druggable protein domains’ are accessed by current drugs, despite estimates of 10
000 genes that define our physical existence. However, this fantastic advance only opened further questions and research directions, as the functions of the majority of human gene products remain unknown. In addition, to unravel the functions of those genes, a major task for chemical biologists will be to discover inhibitors and activators of these gene products, both as chemical probes and potential drug leads (chemical genetics).2 Recent studies have revealed that only 266 human genome-derived targets of the druggable genome (estimated to be ∼3000)3 are currently targeted by small molecules, with up to twice that number if one considers that proteins with 50% identity may display similar pharmacology. Thus, the dire need for discovery of small molecules that can modulate protein function to address human disease cannot be understated. Indeed, a reasonable extension of the central dogma of biology highlights the great importance of small molecules in biological information flow (Fig. 1) given their proven ability to modulate the activity of biological macromolecules (Fig. 1).4 Looking at this gap in knowledge regarding small molecule–protein interactions in a different way, the fact that only 130 ‘privileged druggable protein domains’ are accessed by current drugs, despite estimates of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 folds5 and more than 16
000 folds5 and more than 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 protein families, is staggering.6 These estimates do not include the genomes of problematic microorganisms that also lead to human disease, including bacteria, fungi, and parasites. So, why should natural products be considered a source of small molecules to assist with filling this gap in knowledge? One commonly posited argument, which is well substantiated, invokes the historical and present value of natural products considering currently used pharmaceutical agents.7 However, the great information content of these ‘genetically-encoded small molecules’,8 resulting from their necessary interaction with the biosynthetic enzymes that produce them and their evolutionary optimization for interaction with their biological targets, cannot be overstated. Thus, we should continue to harvest the information offered by natural products and their interactions with various protein folds that wait to be discovered through identification of their cellular targets.
000 protein families, is staggering.6 These estimates do not include the genomes of problematic microorganisms that also lead to human disease, including bacteria, fungi, and parasites. So, why should natural products be considered a source of small molecules to assist with filling this gap in knowledge? One commonly posited argument, which is well substantiated, invokes the historical and present value of natural products considering currently used pharmaceutical agents.7 However, the great information content of these ‘genetically-encoded small molecules’,8 resulting from their necessary interaction with the biosynthetic enzymes that produce them and their evolutionary optimization for interaction with their biological targets, cannot be overstated. Thus, we should continue to harvest the information offered by natural products and their interactions with various protein folds that wait to be discovered through identification of their cellular targets.
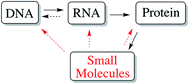 | ||
| Fig. 1 Modified central dogma of biology showing the importance of small molecules, including natural products, and their impact on biological information flow. | ||
Roughly a decade ago, it was suggested that a renaissance in natural products research was coming.9 It may be safe to say that we are now living in the era of a renewed appreciation, with the attendant discovery opportunities for natural products research. The impact on cell biology, enabled through studies of natural products, is broad and continues to increase. Some examples include the nexus of transcriptional control enabled through studies of FK506, cyclosporine, and rapamycin; microtubule dynamics enabled by studies of taxol, epothilone, and discodermolide; G-protein coupled receptors through studies of epinephrine and the cannabinoids; translational control enabled by studies of homoharringtonine, pateamine A, and hippuristanol; ion channels enabled by saxitoxin and several marine polyethers including brevetoxin; proteasomal pathways through studies of proteasome inhibitors including epoxomicin, lactacystin, and salinosporamide; and finally the modification of histones enabled by studies of trapoxin, to name just a few. A great upsurge in the study of natural products is most readily seen in the following areas: (a) biosynthesis and synthetic biology; (b) detailed molecular level studies of natural products through collaborative efforts of synthetic chemists and cell biologists; (c) chemical ecology, leading to hypothesis-driven, natural product discovery. In the post-genomic era of drug discovery, health scientists are redirecting their efforts toward ‘precision medicine’ and ‘personalized medicine’ that takes into account a patient's individual genome. Therefore, a molecular level understanding of natural products and their intended, as well as off targets, is an essential first step in identifying potentially novel druggable targets for treating human disease. These studies have and will continue to provide new platforms for drug discovery.
In this Themed Issue, several contributed articles describe various genome-wide, proteome-wide, phage display, and database mining approaches which compliment more established techniques, e.g. affinity chromatography techniques, for assigning function to natural products.
The Highlight by Yao et al. (DOI: 10.1039/C5NP00101C) discusses advances in photoaffinity labelling through the scalable synthesis of minimal photoactivatable reagents that can be attached to natural products for subsequent studies in live cells. This proteome-wide strategy importantly provides information regarding both high- and low-affinity cellular targets of natural products.
The Review by Sasse, Brönstrup, Prochnow, and Fetz (DOI: 10.1039/C5NP00113G) describes recent advances in image analysis involving both manual and more recent automated approaches. In particular, through direct observation or specific labelling of structural elements and selected proteins in cells with dyes, a comparison of phenotypic effects induced by natural products are directly observable under the microscope. These visualization strategies with whole cells enable direct detection of phenotypic changes induced by natural products and subsequent correlation for identification of both bacterial and eukaryotic cellular targets.
Database mining methods involving MorphobBase and ChemProteoBase are highlighted by Osada, Futamura, and Muroi (DOI: 10.1039/C5NP00106D). With the advent of large data sets derived from profiling methods and perturbations invoked by natural products, correlations are enabling cellular target identification through use of these databases.
Brown and Farha (DOI: 10.1039/C5NP00127G) describe both genomic and biochemical approaches for cellular target identification of antimicrobial natural products. The advent of modern technologies for chemical biology has enabled several new screening platforms for mechanism of action studies applicable to antimicrobial agents that should foster further discovery of novel targets for antibiotics.
Karuso and Piggott (DOI: 10.1039/C5NP00128E) present a tutorial on the application of the T7 bacterial phage display technique to aid in target identification of natural products. The bacterial phage display system was originally developed to display libraries of small peptides on phage surfaces. It was subsequently refined to display proteins. The relatively ease with which to create and screen a phage library displaying a large number of proteins renders this technique a unique and complementary method for target identification.
Sieber and Wright (DOI: 10.1039/C6NP00001K) describe the growing utility of proteomic profiling methods using natural products that are armed with a ‘minimal’ alkyne or azide reporter tag to enable cellular target studies in situ with live cells. The authors cover the use of this strategy in a ‘competitive mode’ format to enable identification of small molecules including other natural products that interact with well-characterized cellular probes for protein families.
Several ‘case studies’ highlight specific examples of synthetic studies or development of specialized techniques to enable a molecular level understanding of the cellular targets of natural products.
In the Highlight by Lei, Li, and Jones (DOI: 10.1039/C5NP00089K), synthetic and mechanistic studies of the oligomeric sesquiterpenoids ainsliadimer A and ainsliatrimer that inhibit NF-kB signaling, leading to potent anti-inflammatory activity, are discussed. In addition, they describe their new bioorthogonal ligation (TQ-ligation) for target identification. The finding of a previously untargeted allosteric site in NF-kB demonstrates the utility of natural products in identifying novel protein folds in well-studied proteins.
Koide and Pham (DOI: 10.1039/C5NP00110B) provide a Highlight that demonstrates the utility of natural products in discovering novel, potential druggable targets, in this case, the spliceosome. In a Highlight by Chen (DOI: 10.1039/C5NP00153F), the importance of ecological observations to the discovery of novel natural products is demonstrated. These studies led to the discovery of the natural product cyclopamine, which was found to be the first specific inhibitor of Hedgehog signaling and an antagonist of Smoothened, a transmembrane receptor.
Kakeya's Highlight (DOI: 10.1039/C5NP00120J) describes their strategies toward understanding the microbial natural products that target cancer through the microenvironment of cancer cells and cell membrane signaling. The development of two technologies for cellular target identification are also described, which could have broad utility.
A few Highlights address the challenge of synthesizing an appropriate cellular probe from a natural product, given that identifying an appropriate attachment point continues to be a bottleneck for subsequent mechanism of action studies. Kanoh's Highlight (DOI: 10.1039/C5NP00117J) describes a unique strategy for immobilizing natural products through direct cross-linking to generate affinity matrices that can be used for whole proteome profiling. The use of photogenerated carbenes that react with natural products enables a randomized strategy for modification of natural products, resulting in a small collection of attachment points. Several examples of how this strategy has been utilized to identify the cellular targets of natural products, demonstrating the potential of this strategy, along with a tutorial for its application, are described. Kwon, Kim and Chang (DOI: 10.1039/C5NP00107B) review the development and application (case studies) of four methods that importantly make use of unmodified natural products: drug affinity responsive target stability (DARTS), stability of proteins from rates of oxidation (SPROX), cellular thermal shift assay (CETSA), and thermal proteome profiling (TPP).
The editors hope this Themed Issue will enable readers to discover the fantastic, enduring, and ever-increasing vibrancy in research focused on target identification of natural products. We anticipate that this research endeavor will continue to lead to new druggable cellular targets and ultimately new treatments for a broad spectrum of human disease in the coming decades. Finally, this Themed Issue would not have been possible were it not for the dedication and hard work of contributing authors and reviewers, so we wish to close with a heartfelt extension of gratitude for their contributions to this issue!
References
- K. Nicolaou and T. Montagnon, Molecules that changed the world, Wiley-VCH, Weinheim, 2008 Search PubMed.
- R. L. Strausberg and S. L. Schreiber, Science, 2003, 300, 294–295 CrossRef CAS PubMed.
- (a) J. P. Overington, B. Al-Lazikani and A. L. Hopkins, Nat. Rev. Drug Discov., 2006, 5, 993–996 CrossRef CAS PubMed; (b) A. L. Hopkins and C. R. Groom, Nat. Rev. Drug Discov., 2002, 1, 727–730 CrossRef CAS PubMed; (c) For the launch of the “Illuminating the Druggable Genome” effort at the National Institutes of Health in 2013, see: M. Griffith, et al. , Nat. Methods, 2013, 10, 1209–1210 CrossRef CAS PubMed.
- Adapted by permission from Prof. Jon Clardy (Harvard Medical School).
- E. V. Koonin, Y. I. Wolf and G. P. Karev, Nature, 2002, 420, 218–223 CrossRef CAS PubMed.
- D. Vitkup, E. Melamud, J. Moult and C. Sander, Nature Struct. Biol., 2001, 8, 559–566 CrossRef CAS PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2012, 75, 311–335 CrossRef CAS PubMed.
- S. J. Miller and J. Clardy, Nat. Chem., 2009, 1, 261–263 CrossRef CAS PubMed.
- (a) I. Paterson and E. A. Anderson, Science, 2005, 310, 451–453 CrossRef PubMed; (b) F. E. Koehn and G. T. Carter, Nat. Rev. Drug Discov., 2005, 4, 206–220 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |
