Open Access Article
Open Access ArticleQuantum dots as a fluorescent labeling tool for live-cell imaging of Leptospira†
Yotsakorn
Tantiapibalkun
a,
Sopon
Nuchpun
 a,
Wid
Mekseriwattana
a,
Wid
Mekseriwattana
 b,
Sukhonta
Limsampan
c,
Galayanee
Doungchawee
d,
Kulachart
Jangpatarapongsa
b,
Sukhonta
Limsampan
c,
Galayanee
Doungchawee
d,
Kulachart
Jangpatarapongsa
 c,
Toemsak
Srikhirin
c,
Toemsak
Srikhirin
 e and
Kanlaya Prapainop
Katewongsa
e and
Kanlaya Prapainop
Katewongsa
 *ae
*ae
aDepartment of Biochemistry, Faculty of Science, Mahidol University, Bangkok, 10400, Thailand. E-mail: kanlaya.pra@mahidol.edu
bDepartment of Chemistry, Faculty of Science, Chulalongkorn University, Bangkok, 10330, Thailand
cCenter for Research Innovation and Biomedical Informatics, Faculty of Medical Technology, Mahidol University, Nakhon Pathom, 73170, Thailand
dDepartment of Pathobiology, Faculty of Science, Mahidol University, Bangkok, 10400, Thailand
eSchool of Materials Science and Innovation, Faculty of Science, Mahidol University, Bangkok, 10400, Thailand
First published on 5th July 2024
Abstract
Leptospirosis is a global public health problem caused by Gram-negative pathogenic bacteria belonging to the genus Leptospira. The disease is transmitted through the urine of infected animals, which contaminates water and soil, leading to the infection of other animals and humans. Currently, several approaches exist to detect these bacteria; however, a new sensitive method for the live-cell imaging of Leptospira is required. In this study, we report the green synthesis of cadmium telluride quantum dots (CdTe QDs) which are unique fluorescent nanocrystals with a high fluorescence quantum yield capable of modifying cell surfaces and are biocompatible with cells. The fabrication of QDs with concanavalin A (ConA), a carbohydrate-binding lectin and known biological probe for Gram-negative bacteria, produced ConA-QDs which can effectively bind on Leptospira and exhibit strong fluorescence under simple fluorescence microscopy, allowing the live-cell imaging of the bacteria. Overall, we performed the simple synthesis of ConA-QDs and demonstrated their potential use as versatile fluorescent probes for the live-cell imaging of Leptospira. This technique could be further applied to track leptospiral cells and study the infection mechanism, contributing to a more thorough understanding of leptospirosis and how to control it in the future.
Introduction
Leptospira is a genus of spirochaetes and causes a disease known as “leptospirosis”. These bacteria can widely spread from infected animals through urine-contaminated water or soil. They can survive for periods ranging from weeks to months in the environment and cause disease transmission through wild or domestic animals. Humans can be infected via direct contact with the urine or other body fluids of infected animals or indirectly through contaminated water, soil, and food. Thus, the risk of contracting leptospirosis is high for many people who work outdoors or practice activities in which they may come in contact with animals, such as fishing, swimming, or hunting, as they could be infected through abrasion of the skin or mucous membranes or from a cut or scratch. Most infected people do not show symptoms;1,2 however, some may experience severe to life-threatening symptoms. The cause of this variability in the manifestation of symptoms is still unknown but could be related to the infection mechanism. Although several detection methods have been extensively developed, there is still limited information and fundamental knowledge regarding the mechanisms of leptospirosis infection. One of the main obstacles is the lack of imaging tools to detect these small-sized bacteria. A common method used for the visualization of Leptospira is dark-field microscopy. However, this technique is not widely available and still has limitations for live-cell tracking. Therefore, a new labeling tool is required. Quantum dots (QDs) are unique fluorescent nanocrystals that have been used in various bioimaging applications.3–5 Combined with concanavalin A (ConA), which is a lectin that binds specifically to polysaccharides, QDs can be used as a detection platform for several targets, including glucose,6Candida albicans,7 or glycan-positive cancer cells, such as those of the MCF7 breast cancer cell line.8 However, the utilization of ConA-QDs for Leptospira labeling has not been reported yet. In the present study, we describe the synthesis of ConA-functionalized QDs and demonstrate their use as a fluorescent labeling tool for the detection of Leptospira. The ConA-QDs developed in this study could be further applied to track leptospiral cells and study the infection mechanism, contributing to a deeper understanding of leptospirosis and how to control it in the future.Experimental section
Materials
Cadmium chloride hemi(pentahydrate), sodium borohydride (NaBH4), mercaptosuccinic acid (MSA), tellurium powder, concanavalin A (ConA) from Canavalia ensiformis (jack bean), sodium dodecyl sulfate, sodium cacodylate trihydrate, osmium tetroxide, and 0.01% poly-L-lysine solution were purchased from Sigma-Aldrich, USA. Methanol was obtained from Loba Chemie, India, while 30% acrylamide/bis solution (29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and tetramethylethylenediamine were purchased from Bio-Rad, USA. Tris hydrochloride and ammonium persulfate were obtained from Vivantis, USA. For immunoblotting, rabbit anti-Shermani antibodies were prepared according to a previously reported method.9 Goat anti-rabbit immunoglobulin-HRP antibodies were purchased from DakoCytomation, UK.
1) and tetramethylethylenediamine were purchased from Bio-Rad, USA. Tris hydrochloride and ammonium persulfate were obtained from Vivantis, USA. For immunoblotting, rabbit anti-Shermani antibodies were prepared according to a previously reported method.9 Goat anti-rabbit immunoglobulin-HRP antibodies were purchased from DakoCytomation, UK.
Synthesis of CdTe QDs
Water-soluble CdTe QDs were prepared via an aqueous synthesis route modified from the protocol described by Ebrahim et al., and the synthesis process is shown in Fig. 1.10 In brief, NaBH4 (80 mg) and Te powder (31 mg) were dissolved in deionized water and stirred at 60 °C under a N2 atmosphere until the solution turned pink, generating a NaHTe solution. The solution was then transferred to a glass syringe and added dropwise to a 100 mL solution of CdCl2 (2.4 mM) and MSA (5.5 mM) (pH preadjusted to 11.2). The mixture was stirred at 90 °C for 1 h. The QDs were precipitated by adding ethanol and collected by centrifugation. The nanoparticles were resuspended in deionized water and stored at 4 °C in darkness.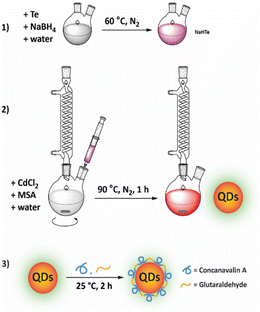 | ||
| Fig. 1 Illustration of the process of QD synthesis. (1) Generating NaHTe solution at 60 °C under a N2 atmosphere. (2) Synthesis of QD-MSA. (3) Fabrication of QDs with ConA. | ||
Coating of QDs with ConA (ConA-QDs)
2 mL of CdTe QDs were coated with a ConA solution at a concentration of 0.3 mg mL−1 and 1% glutaraldehyde. The mixture was gently stirred for 2 h at 25 °C. The resulting ConA-QDs were isolated using centrifugal filters (Mw cut-off = 30 kDa, Amicon® Ultra-15, Merck, USA). The obtained particles were stored at 4 °C until further use.Characterization of the QDs
The hydrodynamic diameter (DH) and zeta potential of the synthesized QDs were measured using the dynamic light scattering method (Zetasizer Nano ZS90, Malvern, UK). Spectroscopic properties were characterized using an Agilent 8453 UV-Vis spectrophotometer and a Shimadzu RF-5301PC spectrofluorophotometer. The core sizes of the QDs were calculated based on their first UV-Vis absorption peak by following eqn (1), as previously reported,11 where λ is the wavelength of the first excitation peak measured in nanometers (nm) and D is the average diameter of the QDs, also in nm. The morphology of the QDs was observed under a transmission electron microscope (TEM, FEI Tecnai T20 G2, Olympus). The amount of ConA on the QDs was determined by performing a micro bicinchoninic acid assay (Micro BCA assay).| D = (9.8127 × 10−7)λ3 − (1.7147 × 10−3)λ2 + (1.0064)λ − (194.84) | (1) |
Leptospira strains and their cultivation
L. santarosai serovar Shermani and a patient-isolated L. interrogans strain of unknown serotype (labeled 53N5) were used in this research. The former strain was provided by the National Institute of Health of Thailand (NIH), while strain 53N5 was isolated from Nan province, Thailand, in 2010. Additional details with regard to this isolate can be found in the Public database for molecular typing and microbial genome diversity (PubMLST) under identification number 599. The Leptospira strains were cultured in Ellinghausen–McCullough–Johnson–Harris medium at 30 °C. The bacteria were maintained and cultivated at weekly intervals or until cell density reached 1 × 108 cells per mL.Biocompatibility of QDs with L. santarosai
L. santarosai was treated with QDs at a final concentration of 100 to 900 nM and incubated for up to 72 h. Samples were collected at time intervals, and cell growth profiles and cell viability were determined by measuring optical density (O.D.) at 420 nm and direct cell counting under dark-field microscopy.Leptospira–ConA agglutination assay
L. santarosai was cultured until reaching a cell density of 1 × 108 cells per mL. Cells were transferred to a 96-well plate and treated with a solution of ConA (2 mg mL−1). The cells were then incubated for 2 h, and agglutination was evaluated under a dark-field microscope. The anti-Shermani specific antibodies were used as positive control.SDS-PAGE and western blot analysis
Samples from the agglutination assay were centrifuged to remove unbound reactants. The pellets were collected, mixed with loading dye, and heated at 95 °C for 5 min. The samples were loaded into 12% polyacrylamide gel and separated at a constant voltage of 120 V for 90 min. Protein patterns were visualized by staining with Coomassie Brilliant Blue R-250.For western blotting, proteins were transferred from the polyacrylamide gel onto a nitrocellulose membrane. Nonspecific bindings were blocked with 5% skim milk, and the membrane was incubated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution of rabbit anti-Shermani antibodies for 1 h at room temperature followed by incubation with anti-rabbit HRP secondary antibodies and the blotting was visualized using DAB as an HRP substrate.
1000 dilution of rabbit anti-Shermani antibodies for 1 h at room temperature followed by incubation with anti-rabbit HRP secondary antibodies and the blotting was visualized using DAB as an HRP substrate.
Labeling of Leptospira with QDs
The Leptospira strains were incubated with either QDs or ConA-QDs at a final concentration of 600 nM for 2 h. The samples were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min to remove excess QDs. The pellets were resuspended in the cultivation media and placed on a glass slide, and the labeled bacteria were observed under a fluorescence microscope (Olympus BX53).
000 rpm for 15 min to remove excess QDs. The pellets were resuspended in the cultivation media and placed on a glass slide, and the labeled bacteria were observed under a fluorescence microscope (Olympus BX53).
The interaction between the QDs and Leptospira was further confirmed by scanning electron microscopy. The resuspended samples were placed onto coverslips and pre-coated with poly-L-lysine. The cells were fixed with glutaraldehyde and treated with osmium reagent. They were then dehydrated through serial concentrations of ethanol solution and subjected to platinum/palladium coating. Finally, the cells were visualized under a HITACHI SU-8010 field-emission scanning electron microscope.
Results and discussion
Characterization of the QDs and ConA-QDs
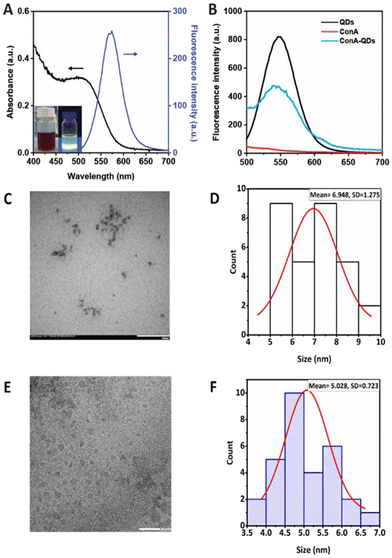
The MSA-capped CdTe QDs successfully obtained from the simple aqueous synthesis appeared as a brown solution under ambient light and exhibited fluorescence properties under UV light, with an absolute quantum yield of 23.15 and a high quantum yield of 86% compared with rhodamine 6G. The QDs showed broad UV-Vis absorbance with a maximum excitation wavelength of 530 nm, enabling a wide range of excitation wavelengths. In addition, the QDs exhibited a narrow emission spectrum with a maximum emission wavelength of 554 nm (Fig. 2A). The full width at half maximum of the peak was estimated at 55 nm, which is considered narrow and thus suitable for biolabeling purposes.12 The approximate core size of the QDs was calculated from the absorption wavelength using eqn (1), as previously reported,11,13 and was determined to be 2.98 nm. The TEM images and the size analysis using ImageJ also supported the size from the calculation (Fig. 2C–F). The QDs had a DH of 5.7 ± 4.9 nm in water (ESI Fig. S1†). However, the QDs were further used in cell culture media. Therefore, the QDs were analyzed under cell culture conditions, which demonstrated a DH of 127.1 ± 1.2 nm, a polydispersity index (PdI) of 0.3, and a zeta potential of −35.20 ± 2.74 mV (Table 1). A larger hydrodynamic size of particles in culture media was observed for other nanoparticles such as silica nanoparticles,14 polymeric nanoparticles,15 and magnetic nanoparticles.16Due to their fluorescence properties, QDs have been used as fluorescent labels in bacteria, such as uncoated CdSe QDs for Escherichia coli (E. coli) detection.17 Modifications of the QD surface increase selectivity, such as modification of silicon QDs with D-alanine leads to Gram-positive bacteria labeling, whereas using gluconamide enhances selectivity to Gram-negative bacteria.18 In addition, QDs functionalized with RBP 55, a phage receptor binding protein, show increased sensitivity and specificity in Salmonella detection.19
In this work, ConA was employed as a coating agent to enhance the interaction of QDs with Leptospira. ConA is a well-established lectin binding to α-mannosyl and glucosyl of polysaccharides, and can interact with lipopolysaccharides on the surface of Gram-negative bacteria. For example, it has been used to conjugate with Rose Bengal to increase the antimicrobial properties against Gram-negative bacteria in photodynamic therapy.20 It was coated on magnetic nanoparticles to increase the electrochemical response for bacteria detection.21 Even though ConA is widely used, there is no report on using ConA to enhance the fluorescence labeling properties of QDs for Leptospira detection. ConA was crosslinked through reaction with glutaraldehyde and attached to the QD surface via electrostatic interaction, as evidenced by the slight increase in zeta potential to −32.00 ± 0.95 mV following the coating process. After coating the QDs with ConA to obtain ConA-QDs, these exhibited a broader fluorescence emission spectrum with lower intensity than the pristine QDs, which was expected due to the larger distribution of nanoparticle sizes and energy transfer between the QDs and ConA (Fig. 2B).10 The size of the ConA-QDs increased, as shown by the observed DH value of 860.2 ± 38.8 nm with a PdI of 0.6 (Table 1). The increased size was due to the extra layers of ConA on the surfaces, which acted as bridges allowing interparticle crosslinking.
Biocompatibility of QDs with L. santarosai
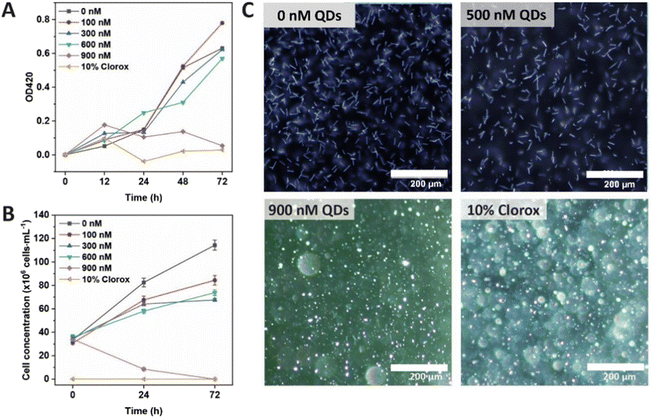
As Cd-containing QDs are known to be toxic for live cells,22,23 their effect on the growth of leptospiral cells was tested prior to biolabeling. In this study, L. santarosai serovar Shermani, the most predominant serovar found in many countries, including Thailand24 and Taiwan,25 was chosen as a model. QDs at different concentrations (0–900 nM) were incubated with L. santarosai for 72 h. The biological effects of QDs were evaluated by measuring the O.D. at 420 nm, which is a common method to determine bacterial viability,26,27 and dark-field microscopy was used to visualize and count the cells. No obvious reduction in cell viability was observed for Leptospira when the strains were exposed to QDs at concentrations of up to 600 nM throughout the entire 72 h incubation time (Fig. 3A). Conversely, when the QD concentration was increased to 900 nM, cell viability drastically dropped after 24 h, and a similar viability was observed in the negative control group treated with bleach at the end of the experiment.To validate the O.D. measurements, bacteria were also observed under a dark-field microscope to directly count the number of viable cells (Fig. 3B). The results were consistent with the cell viability curve obtained from the O.D. measurements. In addition, the dark-field images showed that the cells were still intact and similar in terms of morphology (rod shape) to those treated with distilled water (Fig. 3C and Fig. S2, ESI videos 3–9†). Therefore, the QDs could be applied to Leptospira at concentrations of up to 600 nM for 72 h. A comparison between L. santarosai treated with 500 nM and 900 nM QDs is shown in Fig. 3C. In addition, the Leptospira that were treated with 600 nM QDs for 72 h can be subcultured (Fig. S3†). The motility was the same as that of the control with no sign of toxicity (ESI video 10†).
Biolabeling of leptospiral cells with ConA-QDs
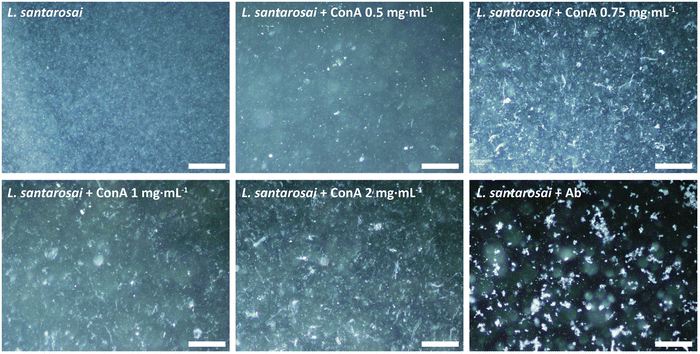
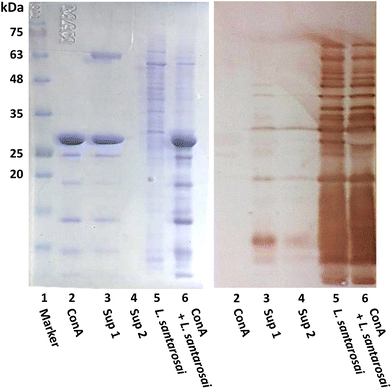
ConA is known to interact with α-mannosyl and glucosyl residues of polysaccharides which are highly expressed on Gram-negative bacteria,28 and it was expected to bind to the surface of leptospiral cells, forming agglutinates. To confirm this prediction, an agglutination assay was performed on L. santarosai and observed under a dark-field microscope, and positive results were obtained. The spirochete formed agglutinates in a dose-dependent manner and, specifically, formed the largest ones when incubated with ConA at a concentration of 2 mg mL−1 (Fig. 4). Therefore, this ConA concentration was used for further analysis. This agglutinate formation was similarly observed when Leptospira interacted with specific anti-Shermani antibodies.The interaction was further confirmed by SDS-PAGE and western blotting (Fig. 5). After incubation with ConA, unbound ConA was successfully removed, as shown in lane 4 (Fig. 5), where no protein bands are present in the second supernatant. The results showed the coexistence of Shermani protein bands and a ConA band at approximately 29 kDa (Fig. 5A). In further analyses using western blotting, ConA proteins showed no cross-reaction with the anti-Shermani antibodies (Fig. 5B, lane 2). The proteins from Shermani alone and a mixture of Shermani and ConA (Fig. 5, lanes 5 and 6) exhibited similar patterns. The intense band at 29 kDa was not present following immunostaining, which verified the origin of the band as ConA protein, suggesting an interaction between Shermani and ConA. These results were expected; ConA could bind with leptospiral cells, possibly by interacting with the rich lipopolysaccharides present on their outer membrane.29
To investigate the potential use of ConA-QDs for Leptospira labeling, L. santarosai was incubated with different concentrations of ConA-QDs (300–500 nM) and observed under a dark-field microscope (Fig. 6). The leptospiral cells incubated with either QD or ConA-QDs showed no differences in terms of morphology. The ConA-QDs did not cause cell toxicity (Fig. S4†). However, when the Leptospira was observed under a fluorescence microscope at excitation wavelengths between 460 and 495 nm, it was shown that the Leptospira labeled with ConA-QDs showed stronger fluorescence than the Leptospira incubated with QDs (Fig. 7). In addition, the fluorescence was much higher at higher concentrations than at lower concentrations of QDs. These results suggested that ConA could enhance the fluorescence labeling of QDs to Leptospira in a dose-dependent manner.
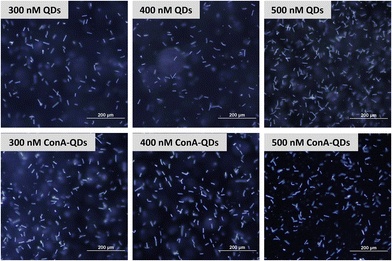 | ||
| Fig. 6 Dark-field microscopy images of L. Shermani incubated with CdTe QDs and ConA-QDs at final concentrations of 300–500 nM. Scale bar = 200 μm. | ||
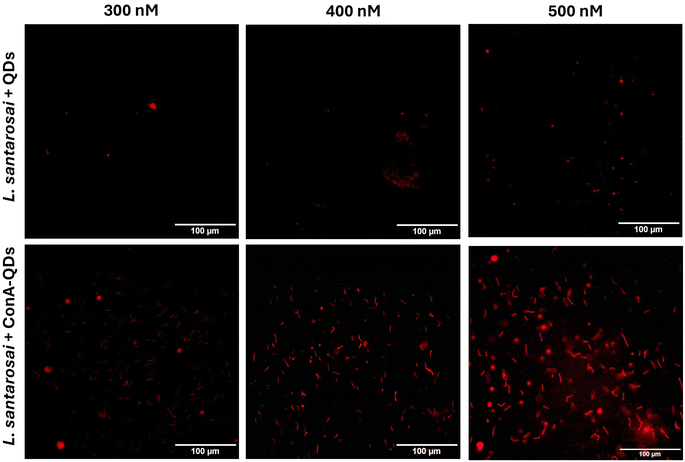 | ||
| Fig. 7 Fluorescence images of L. santarosai incubated with QDs and ConA-QDs at final concentrations of 300–500 nM. Scale bar = 100 μM. | ||
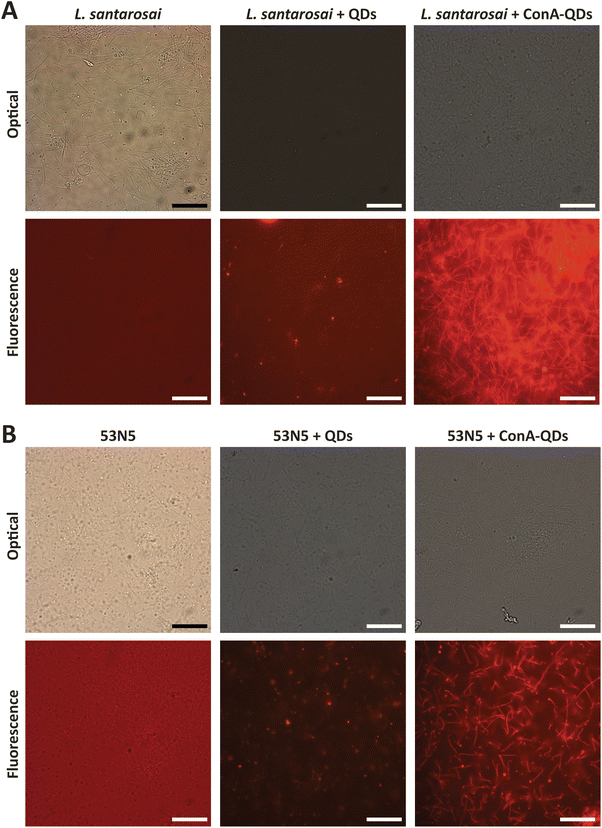
In addition, because fluorescence is a unique property of ConA-QDs as biolabeling tools for Leptospira, two sets of spirochaetes were incubated with either QDs or ConA-QDs for 2 h and observed under a fluorescence microscope (Fig. 8). The two species used were L. santarosai and a patient-isolated L. interrogans strain of unknown serotype, labeled 53N5. Without QD labeling, the bacterial cells were difficult to visualize. When the Leptospira strains were incubated with QDs, the nonspecific interaction with them produced a weak fluorescence signal. In contrast, when the strains were incubated with ConA-QDs, a strong fluorescence signal was detected, as illustrated in Fig. 8 and ESI videos 1 and 2.† To emphasize, ESI videos clearly demonstrated the clearer visualization of Leptospira through strong fluorescence. This was due to the strong interaction between the ConA coating and the outer membrane of the bacterial cells, as discussed earlier. This fluorescence labeling is dose-dependent, as we observed less fluorescence when using lower concentrations (Fig. 7 and ESI videos 11 and 12†). There are several other approaches using fluorescence properties for tracking Leptospira to overcome the limitations of commonly used dark field microscopy, in particular the difficulty in visualizing their dynamic changes; for example, the establishment of bioluminescent Leptospira by constructing and producing bioluminescent L. interrogans isolates expressing green fluorescent protein (GFP), monomeric red fluorescent protein (mRFP),30 or luciferase.31,32 Another approach is using synthetic fluorescent bi-arsenical compounds to label the membrane protein expressing tetracysteine of Leptospira for live cell imaging; however, the wild type Leptospira could not be detected by this method.33 In contrast, the present study does not require manipulation of Leptospira. It is worth noting that this interaction was observed not only in the Shermani serovar but also in the human isolated strain 53N5, suggesting that ConA-QDs could be applied for rapid and versatile detection of other Leptospira serovars.
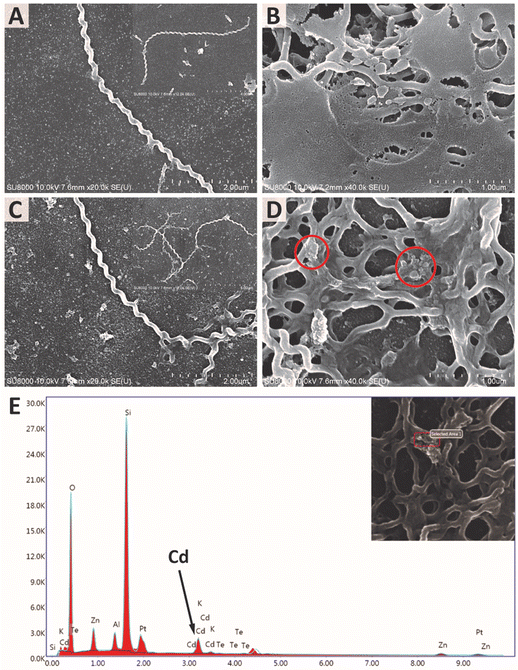
SEM images of both L. santarosai and L. interrogans (53N5) incubated with ConA-QDs were taken to observe the interaction between particles and cells (Fig. 9A–D). In both strains, agglomerates of particles were visible on the surface of bacterial cells (red circles). The large size of the agglomerates suggested that they were not the core QDs but were rather aggregates of the QDs bound together through the interaction between ConA coatings, which in turn interacted with the membranes of leptospiral cells. Fig. 9E shows the EDX analysis of 53N5 cells incubated with ConA-QDs. Further examination of the area with the observed agglomerate showed a signal matching that of Cd, which was the main element of the ConA-QDs used in this study. Therefore, the attachment of ConA-QDs to the surface of Leptospira cells was confirmed.
Conclusion
To the best of our knowledge, this is the first study demonstrating the use of CdTe QDs as biolabeling tools for the imaging of Leptospira. The interaction between the bacterial cells and QDs was enhanced by the application of ConA, and the method showed excellent results with L. santarosai and L. interrogans. This method provides fast imaging of pathogens with high image contrast. The use of QDs as labeling tools allows a wide range of excitation wavelengths, thus minimizing the limitations of imaging equipment. Being characterized by enhanced interaction and excellent fluorescence properties, ConA-QDs are suitable for further development as rapid detection platforms for Leptospira.Author contributions
Y. T. and S. N. designed and performed all the experimental work related to the synthesis of quantum dots, performed in vitro studies, analyzed data, and prepared the original draft. W. M. assisted in the synthesis and characterization of quantum dots, prepared an original draft, and reviewed and edited the manuscript. S. L. helped to obtain dark-field microscopy images. G. D. helped in research design, supported laboratory equipment and specimens, acquired funding, and prepared the manuscript (original draft, reviewed and edited). K. J. supported specimens and laboratory equipment. T. S. helped in the research design and revised the manuscript. K. P. K. designed and supervised the overall research and data analysis, provided criticism, prepared the manuscript (original draft, reviewed and edited), supported laboratory equipment and specimens and acquired funding. All authors have approved the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This research project was supported by Mahidol University under the New Discovery and Frontier Research Grant (2562, NDFR 08/2563 and NDFR 19/2564). This research project has been funded by Mahidol University (Fundamental Fund: fiscal year 2024 by National Science Research and Innovation Fund (NSRF) (FF-079/2567)). Partial support was also provided by a grant from the Center for Scientific Instrumentation and Platform Services, Faculty of Science, Mahidol University. Y. T. and W. M. were supported by the Science Achievement Scholarship of Thailand. S. N. was supported by the Development and Promotion of Science and Technology Talents Project. The authors would like to thank Ms Tasanee Inwisai for her help with cell culture.References
- Leptospirosis, https://www.cdc.gov/leptospirosis/pdf/fact-sheet.pdf.
- P. N. Levett, Clin. Microbiol. Rev., 2001, 14, 296–326 CrossRef CAS PubMed.
- A. M. Wagner, J. M. Knipe, G. Orive and N. A. Peppas, Acta Biomater., 2019, 94, 44–63 CrossRef CAS PubMed.
- M. A. Cotta, ACS Appl. Nano Mater., 2020, 3, 4920–4924 CrossRef CAS.
- R. Bilan, F. Fleury, I. Nabiev and A. Sukhanova, Bioconjugate Chem., 2015, 26, 609–624 CrossRef CAS PubMed.
- J.-H. Wang, Y.-Q. Li, H.-L. Zhang, H.-Q. Wang, S. Lin, J. Chen, Y.-D. Zhao and Q.-M. Luo, Colloids Surf., A, 2010, 364, 82–86 CrossRef CAS.
- D. P. L. A. Tenório, C. G. Andrade, P. E. Cabral Filho, C. P. Sabino, I. T. Kato, L. B. Carvalho, S. Alves, M. S. Ribeiro, A. Fontes and B. S. Santos, J. Photochem. Photobiol., B, 2015, 142, 237–243 CrossRef PubMed.
- S. Jafarzadeh, N. Bargahi, H. B. Shamloo and J. Soleymani, RSC Adv., 2022, 12, 8492–8501 RSC.
- G. Doungchawee, W. Sirawaraporn, A. Icksang-Ko, S. Kongtim, P. Naigowit and V. Thongboonkerd, J. Med. Microbiol., 2007, 56, 587–592 CrossRef CAS PubMed.
- S. Ebrahim, M. Reda, A. Hussien and D. Zayed, Spectrochim. Acta, Part A, 2015, 150, 212–219 CrossRef CAS PubMed.
- W. W. Yu, L. Qu, W. Guo and X. Peng, Chem. Mater., 2003, 15, 2854–2860 CrossRef CAS.
- B. Liu, B. Jiang, Z. P. Zheng and T. C. Liu, J. Lumin., 2019, 209, 61–68 CrossRef CAS.
- O. Thepmanee, K. Prapainop, O. Noppha, N. Rattanawimanwong, W. Siangproh, O. Chailapakul and K. Songsrirote, Anal. Methods, 2020, 12, 2718–2726 RSC.
- K. Prapainop, R. Miao, C. Åberg, A. Salvati and K. A. Dawson, Nanoscale, 2017, 9, 11261–11268 RSC.
- W. Mekseriwattana, A. Phungsom, K. Sawasdee, P. Wongwienkham, C. Kuhakarn, P. Chaiyen and K. P. Katewongsa, Photochem. Photobiol., 2021, 97, 1548–1557 CrossRef CAS PubMed.
- W. Mekseriwattana, T. Thiangtrongjit, O. Reamtong, P. Wongtrakoongate and K. P. Katewongsa, ACS Omega, 2022, 7, 37589–37599 CrossRef CAS PubMed.
- E. Mohamadi, M. Moghaddasi, A. Farahbakhsh and A. Kazemi, J. Photochem. Photobiol., B, 2017, 174, 291–297 CrossRef CAS PubMed.
- J. Lin, L. Xu, Y. Zheng, D. Wu and J. Yue, Front. Bioeng. Biotechnol., 2022, 10, 971682 CrossRef PubMed.
- Y. Ding, W. Zhu, C. Huang, Y. Zhang, J. Wang and X. Wang, Food Chem., 2023, 428, 136724 CrossRef CAS PubMed.
- A. Cantelli, F. Piro, P. Pecchini, M. Di Giosia, A. Danielli and M. Calvaresi, J. Photochem. Photobiol., B, 2020, 206, 111852 CrossRef CAS PubMed.
- A. G. da Silva Junior, I. A. M. Frias, R. G. Lima-Neto, S. R. Sá, M. D. L. Oliveira and C. A. S. Andrade, Microbiol. Res., 2021, 251, 126834 CrossRef CAS PubMed.
- T. Zhang, Y. Hu, M. Tang, L. Kong, J. Ying, T. Wu, Y. Xue and Y. Pu, Int. J. Mol. Sci., 2015, 16, 23279–23299 CrossRef CAS PubMed.
- M. Wang, J. Wang, H. Sun, S. Han, S. Feng, L. Shi, P. Meng, J. Li, P. Huang and Z. Sun, Int. J. Nanomed., 2016, 11, 2319–2328 CAS.
- S. Chadsuthi, D. J. Bicout, A. Wiratsudakul, D. Suwancharoen, W. Petkanchanapong, C. Modchang, W. Triampo, P. Ratanakorn and K. Chalvet-Monfray, PLoS Neglected Trop. Dis., 2017, 11, e0005228 CrossRef PubMed.
- L. F. Chou, T. W. Chen, Y. C. Ko, M. J. Pan, Y. C. Tian, C. H. Chiu, P. Tang, C. C. Hung and C. W. Yang, Emerging Microbes Infect., 2014, 3, e82 CAS.
- S. Schreier, W. Triampo, G. Doungchawee, D. Triampo and S. Chadsuthi, Biol. Res., 2009, 42, 5–12 Search PubMed.
- C. Zavala-Alvarado, S. G. Huete, O. Sismeiro, R. Legendre, H. Varet, G. Bussotti, C. Lorioux, P. Lechat, J. Y. Coppée, F. J. Veyrier, M. Picardeau and N. Benaroudj, PLoS Pathog., 2021, 17, e1009087 CrossRef CAS PubMed.
- A. M. Wu, E. Lisowska, M. Duk and Z. Yang, Glycoconjugate J., 2009, 26, 899–913 CrossRef CAS PubMed.
- S. H. Hsu, C. C. Hung, M. Y. Chang, Y. C. Ko, H. Y. Yang, H. H. Hsu, Y. C. Tian, L. F. Chou, R. L. Pan, F. G. Tseng and C. W. Yang, Sci. Rep., 2017, 7, 8363 CrossRef PubMed.
- F. Aviat, L. Slamti, G. M. Cerqueira, K. Lourdault and M. Picardeau, Appl. Environ. Microbiol., 2010, 76, 8135–8142 CrossRef CAS PubMed.
- G. L. Murray, A. M. King, A. Srikram, R. W. Sermswan and B. Adler, J. Clin. Microbiol., 2010, 48, 2037–2042 CrossRef CAS PubMed.
- G. Ratet, F. J. Veyrier, M. Fanton, d'Andon, X. Kammerscheit, M.-A. Nicola, M. Picardeau, I. G. Boneca and C. Werts, PLoS Neglected Trop. Dis., 2014, 8, e3359 CrossRef PubMed.
- C. Hillman, P. E. Stewart, M. Strnad, H. Stone, T. Starr, A. Carmody, T. J. Evans, V. Carracoi, J. Wachter and P. A. Rosa, Front. Cell. Infect. Microbiol., 2019, 9, 287 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Size of QDs in water. Dark-field images of Shermani cells incubated with 100 nM, 300 nM, or 600 nM QDs for 0–72 h. Subculture after treatment with QDs for 72 h. Cell growth of Leptospira after incubation with ConA-QDs. Video 1: L. santarosai after incubation with ConA-QDs under a fluorescence microscope. Video 2: L. interrogans (53N5) after incubation with ConA-QDs under a fluorescence microscope. Videos 3–10: Dark field microscopy of Shermani cells incubated with QDs. Videos 11and 12: Fluorescence of Shermani cells with 300 and 400 nM ConA-QDs. See DOI: https://doi.org/10.1039/d4nr00543k |
| This journal is © The Royal Society of Chemistry 2024 |