Open Access Article
Open Access ArticleFew-layered graphene increases the response of nociceptive neurons to irritant stimuli†
Lieselot
Deleye‡
a,
Francesca
Franchi‡
ab,
Martina
Trevisani
ac,
Fabrizio
Loiacono
 b,
Silvia
Vercellino
b,
Silvia
Vercellino
 ab,
Doriana
Debellis
d,
Nara
Liessi
e,
Andrea
Armirotti
e,
Ester
Vázquez
ab,
Doriana
Debellis
d,
Nara
Liessi
e,
Andrea
Armirotti
e,
Ester
Vázquez
 f,
Pierluigi
Valente§
f,
Pierluigi
Valente§
 *bc,
Valentina
Castagnola§
*bc,
Valentina
Castagnola§
 *ab and
Fabio
Benfenati§
*ab and
Fabio
Benfenati§
 *ab
*ab
aCenter for Synaptic Neuroscience and Technology, Istituto Italiano di Tecnologia (IIT), Largo Rosanna Benzi 10, 16132 Genova, Italy. E-mail: valentina.castagnola@iit.it; fabio.benfenati@iit.it
bIRCCS Ospedale Policlinico San Martino, Largo Rosanna Benzi 10, 16132 Genova, Italy
cDepartment of Experimental Medicine, Section of Physiology, University of Genova, Genoa, 16132, Italy. E-mail: pierluigi.valente@unige.it
dElectron Microscopy Facility, IIT, Via Morego 30, 16163, Genoa, Italy
eAnalytical Chemistry Facility, IIT, via Morego, 30, 16163, Genoa, Italy
fFacultad de Ciencias Químicas, Universidad Castilla La-Mancha, Ciudad Real, 13071 Spain
First published on 9th January 2024
Abstract
The unique properties of few-layered graphene (FLG) make it interesting for a variety of applications, including biomedical applications, such as tissue engineering and drug delivery. Although different studies focus on applications in the central nervous system, its interaction with the peripheral nervous system has been so far overlooked. Here, we investigated the effects of exposure to colloidal dispersions of FLG on the sensory neurons of the rat dorsal root ganglia (DRG). We found that the FLG flakes were actively internalized by sensory neurons, accumulated in large intracellular vesicles, and possibly degraded over time, without major toxicological concerns, as neuronal viability, morphology, protein content, and basic electrical properties of DRG neurons were preserved. Interestingly, in our electrophysiological investigation under noxious stimuli, we observed an increased functional response upon FLG treatment of the nociceptive subpopulation of DRG neurons in response to irritants specific for chemoreceptors TRPV1 and TRPA1. The observed effects of FLG on DRG neurons may open-up novel opportunities for applications of these materials in specific disease models.
Introduction
Since its discovery in 2004, remarkable efforts have been devoted to better understanding the physical and chemical properties of graphene-related materials (GRMs).1–3 This has caused exponential growth in the applications of GRMs in a wide range of fields, from engineering to electronics to biotechnologies and biomedical applications.2–8 GRMs are particularly interesting in neuroscience, as they could overcome the limitations of the currently used materials, such as metals and silicon, which are characterized by elevated stiffness, high inflammatory potential, and poor long-term stability under physiological conditions.9–13 On the contrary, GRMs are characterized by a large surface area, flexibility, and excellent electrical and conductive properties.3,9,14 They could therefore find applications as drug/gene carriers for improving imaging in the brain and as a substrate for tissue engineering.14–22 In this view, GRMs biocompatibility with the chosen biological environment is of great importance. However, in the literature, there is controversial information about the general biocompatibility of GRMs, and it seems that their behavior might be dependent on the specific cellular model employed.23,24 In particular, the results on neurotoxicity are contradictory as graphene has been proven to be toxic, harmless, or even beneficial to the neurons.9,22,25–35 The type of GRM, the dose, functionalization, route of administration, and targeted cells are all factors that strongly contribute to the GRM's biological interactions.In the central nervous system (CNS), many studies predominantly used graphene oxide (GO) or reduced GO (rGO).36–40 There use for applications in the CNS seems promising, but their biocompatibility is still a topic for debate.9,30,31,41–43 Instead, low concentrations of small colloidal few-layered graphene (FLG) flakes showed great biocompatibility towards hippocampal neurons, astrocytes, and blood–brain barrier (BBB).30,31,44
Toxicity studies of GRMs on neurons have mainly focused on the CNS.9,30,31,45,46 Most of the information about GRMs effects on the peripheral nervous system (PNS) refers to planar graphene-based substrates, intended as scaffolds for nerve regeneration.19,21,47–52 Differently from the CNS, peripheral neurons and nerves lack a barrier protecting their neurons from the systemic circulation, and colloidal forms of graphene (also exfoliated from planar implants), purposely or accidentally present in the circulation, might enter in contact with the cells of the PNS. In addition, colloidal GRMs are so far scarcely investigated for their interactions with peripheral neurons.
Somatic sensory neurons, also called dorsal root ganglion (DRG) neurons, have very distinct molecular, morphological, electrical, and network properties compared to central neurons. These cells are the first-order neurons in the somatosensory afferent pathway, transducing peripheral stimuli into electrical information and conveying them to the CNS so they can be analyzed by the brain. Moreover, DRG neurons are essential regulators of pain, and pain-related disorders are known to impact human life quality significantly. GRMs might also purposely be applied to the PNS as, for instance, an analgesic drug carrier for neuropathic pain or direct neuronal regulators, and some research indeed focused on the pain-relieving effects of graphene but did not investigate its direct impact on the sensory neurons’ functionality.53,54
In the present study, colloidal FLG was tested against primary sensory neurons to evaluate its biocompatibility using a palette of readouts, including viability, morphological changes, protein dysregulation, and electrophysiological properties. Given the polymodal nature of DRG neurons mediating diverse stimuli, in this study, we also investigated the response of these neurons to specific modalities evaluating the response to acidic pH and irritants. In this context, we tested capsaicin and allyl isothiocyanate (AITC) that selectively activate two members of the transient receptor potential (TRP) family of ion channels, TRPV1 (vanilloid) and TRPA1 (ankyrin), respectively. We found that DRG neurons, previously incubated with FLG, exhibited an increased response to both irritants, but not to mild acidic pH, a general stimulus for nociceptors. Overall, our results define the safe conditions for the use of colloidal FLG on DRG neurons and pave the way to intriguing new potential applications.
Results and discussion
Primary sensory neurons dissected from P2/P3 rat pups were used as an experimental model to assess the impact of colloidal FLG exposure. DRG cells have distinctive large, round cell bodies that protrude from the planar plane, as shown in Fig. S1A.† Under our culturing conditions, small dividing glial cells were present and necessary for the initial DRG neuron growth.55–57 After one day in vitro (DIV1), the antimitotic cytosine β-D-arabinoside was added to attenuate the growth of these glial cells (Fig. S1B†). At DIV4, neurons were incubated with FLG flakes at concentrations ranging from 10 to 100 μg mL−1. The characterization of our FLG is reported in our previous works.58 One day later, we investigated FLG uptake, neuron viability, presence of apoptosis, alterations in the DRG neuron morphology, protein content, and electrical activity and characteristics.FLG is actively internalized by DRG neurons
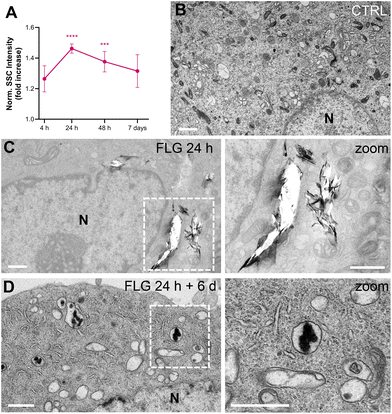
After DRG incubation with FLG, the presence of large flakes outside the cells was observed by optical and scanning electron microscopy (Fig. S2†). The actual internalization of FLG flakes was assessed using flow cytometry, confocal microscopy, and transmission electron microscopy (TEM).Flow cytometry allows for quantification of graphene uptake in a well-established label-free setting,59–61 as a function of the increased cells’ granularity, which in turn increases the measured side scattering (SSC).62 We chose not to label FLG flakes, thus avoiding any possible bias in the graphene–neuron interaction, and measured the uptake kinetics of FLG by SSC values normalized over untreated control samples (CTRL, Fig. 1A). The SSC signal for FLG-treated neurons started increasing after 4 h incubation and reached a plateau of significant internalization after 24 h. From 48 h of incubation up to 7 days, the quantified uptake slightly decreased. This might be due to the material that, after the first 24 h, reaches a maximum of internalization, after which it becomes too aggregated in the medium to be further internalized. Also, degradation or export phenomena cannot be excluded.
Both confocal imaging and TEM were used to visualize the presence of graphene flakes inside the neurons. Graphene flakes can be visualized under a confocal microscope with mid-infrared reflective light; however, quantification through this methodology is difficult as smaller flakes might be undetectable, and some signals from the intracellular compartments might interfere.63 From a qualitative point of view, FLG was localized intracellularly (Fig. S3†). TEM imaging confirmed this observation. In the TEM micrographs shown in Fig. 1, some FLG accumulation in intracellular vesicles is visible after 24 h of exposure (Fig. 1C) compared to untreated cells (Fig. 1B). As can be noticed from Fig. 1C, the FLG flakes appear in large aggregates inside the cell, making it difficult to reach a proper contrast for the images (in comparison to Fig. 1B control, where all the intracellular organelles are clearly stained) and originating some holes during the slicing procedure. The presence of large agglomerates of FLG is in line with the low dispersibility of FLG in aqueous-based media.44
We also observed the FLG flakes’ morphology after being processed by the cells for extended periods. We incubated DRG neurons with 10 μg mL−1 of FLG for 24 h (representing the condition for maximum uptake). After replacing the medium and six further days of culture, cells were observed by TEM (Fig. 1D). Under these conditions, almost no holes were produced by the slicing procedure, and less internalized material was visible. In addition, the morphology appeared quite different, from flake-like after 24 h to amorphous carbon after 6 further days of cell processing. This might suggest the occurrence of some degradation phenomena, although data are not sufficient to conclude it definitively.
Biocompatibility of FLG on DRG neurons
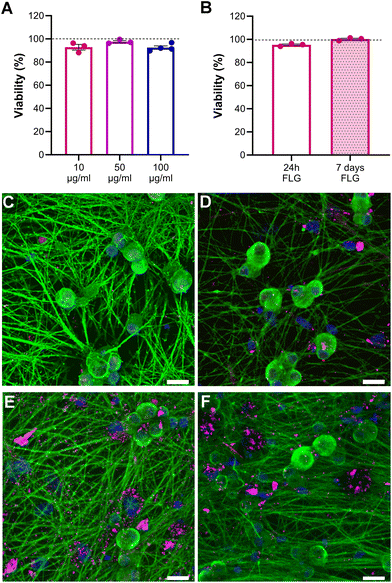
Neuron viability upon FLG incubation at different concentrations was assessed through live calcein/propidium iodide (PI) staining to visualize both viable and dead neurons. Our data showed that the neuronal viability of FLG-treated neurons with respect to untreated neurons cultured under the very same conditions remained unchanged, even at high concentrations of FLG (Fig. 2A; Fig. S4†). Caspase3/7 staining was also performed to visualize the presence of apoptotic cells and to avoid an overestimation of neuron viability. The percentage of caspase-positive neurons remained below 2% under all experimental conditions, indicating the absence of apoptosis after FLG treatment (Fig. S5A–C†). Overall, no signs of cytotoxicity were observed after FLG exposure.We also investigated the effects of prolonged contact with the lowest FLG concentration by assessing cell viability after 7 days (DIV11) of continuous GRM exposure. As can be observed in Fig. 2B, the viability after prolonged FLG exposure was not impaired over time, rather it slightly increased. This can be explained by the fact that DRG neurons tend to cluster in long-term cultures (Fig. S6A–D†) because of their intrinsic tendency to form a ganglion structure, becoming more resistant to the external environment. Therefore, after 7 days, the DRG neuron density in the selected fields was increased if compared to 24 h cultures (Fig. S6E†), resulting in a higher ratio of live over dead cells. These factors can explain the slight increase in viability observed after long-term exposure. Overall, under our experimental conditions, there are no particular concerns in the case of prolonged contact of DRG neurons to FLG.
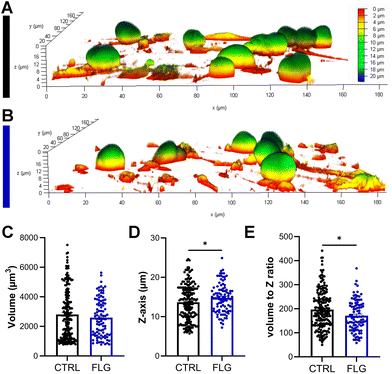
There has been some evidence that the 3D structure of DRG neurons might be affected when they are under stressful conditions.64,65 From optical observations, the neuronal morphology did not seem to be greatly affected after exposure to either concentration of FLG (Fig. 2C–E; Fig. S4†), as previously observed in SEM imaging (Fig. S2†). Confocal z-stack images were then acquired, allowing for the 3D reconstruction and calculation of the soma volume. It must be specified that, to perform this analysis, we selected DRG somas quite isolated from other cells. After exposure to either concentration of FLG, the cell body volume remained substantially unchanged (Fig. 3C; Fig. S7†). However, the shape of the soma was slightly affected after treatment with 100 μg mL−1 of FLG (Fig. 3A and B), with a significant increase in the average Z-length (Fig. 3D). To verify if most neurons with longer Z-lengths were, in general, larger, we calculated the cell volume to Z-length ratio. This ratio was significantly lower after FLG treatment, indicating that the effect was attributable to the change in neuron height (Fig. 3E). These morphological effects were concentration-dependent since the changes were only observed at high concentrations of FLG (Fig. S7†). The change in shape, but not in volume, might indicate an increase in soma elasticity, previously shown in DRG neurons suffering from sciatic nerve injury.64 More research is needed to understand the basis of this phenomenon and the possible connection with cellular stress.
In Fig. S8,† we report a size distribution analysis performed on the diameters of the somas from 2D bright field (BF) images. From the analysis, it can be noticed that in untreated neurons, two main subpopulations can be identified, namely small/medium neurons (15–40 μm diameter) and medium/large (40–60 μm diameter). The size distribution differs from untreated controls when cells are exposed to increasing concentrations of FLG (Fig. S8B and C†). However, given the flakes’ tendency to agglomerate over the cells (see Fig. S8A†), some of the smallest neurons might be lost in the analysis, being covered by the large black spot. Therefore, a significant effect of FLG on neuronal size and morphology cannot be fully demonstrated.
The internalization of FLG in DRG neurons did not show important signs based on the readouts analyzed so far. However, FLG might affect distinct intracellular pathways, resulting in functional changes. Therefore, proteomic analysis was performed on protein lysates from DRG neurons treated for 24 h with either 10 or 100 μg mL−1 of FLG (Fig. 4A and B). However, PCA analysis of the proteomic data did not show a clear separation between CTRL and FLG clusters at both FLG concentrations (Fig. 4A).
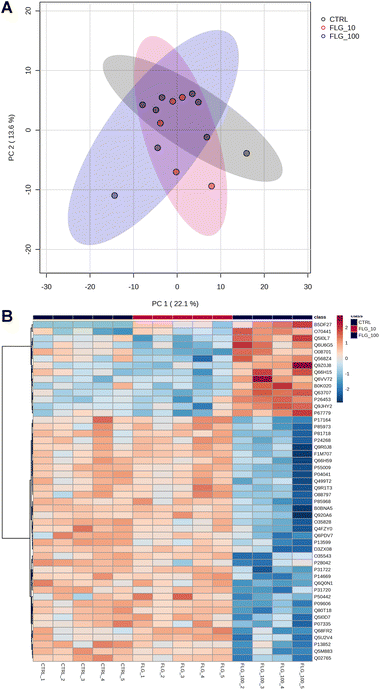 | ||
| Fig. 4 Proteomics analysis. (A) PCA graph of CTRL vs. FLG-treated neurons at the concentrations of 10 and 100 μg mL−1. (B) Heatmap of the 50 most significant proteins following ANOVA test. | ||
Only for the high FLG concentration (100 μg mL−1), the heatmap analysis reveals some significant changes in the protein expression (Fig. 4B). The figure shows the 50 most dysregulated proteins in FLG-treated DRG neurons as compared with CTRL neurons, 14 of which are significantly altered (ANOVA test, corrected p-value <0.05, see ESI files 1 and 2†). Gene ontology analysis indicates that this data are consistent with alterations in glutamate biosynthesis, collagen, and complement pathways (see ESI file 3†).
FLG does not affect the passive and active properties of DRG neurons
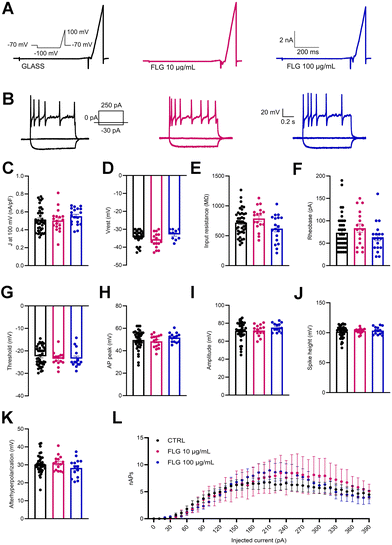
Besides viability, the passive and active electrical properties of DRG neurons are a very sensitive indicator of neuron health and function. In this context, we analyzed the single-cell neuronal properties of control DRG neurons and DRG neurons treated with low (10 μg mL−1) and high (100 μg mL−1) concentrations of FLG by measuring the whole-cell macroscopic currents (Fig. 5A). No significant differences were observed between the two groups in the outward current density (J; nA pF−1) measured at 100 mV (Fig. 5C) using a ramp paradigm.Patch-clamp recordings in current-clamp configuration were used to dissect the firing properties of DRG neurons in the experimental groups under study. To do this, we delivered constant current pulses (1 s duration) of increasing amplitude (in 10 pA steps) to evoke action potentials (APs) (Fig. 5B). Under these conditions, the initial resting membrane potential of treated and untreated DRG neurons was unchanged (Fig. 5D). Moreover, also the rheobase and the input resistance remained substantially unaffected by the FLG treatment (Fig. 5E and F).
To study in-depth the biophysical properties of the single AP, the plot of the time derivative of voltage (dV/dt) vs. voltage was constructed (phase-plot analysis).66,67 The plot was obtained from the first AP elicited by the minimal current injection (rheobase) by current-clamp family step protocol (see Fig. 5B, inset). The following parameters were studied: threshold, AP peak, amplitude, and spike height. As seen in Fig. 5G–J, the presence of either 10 or 100 μg mL−1 FLG did not significantly affect these parameters of AP shape. Finally, the number of APs evoked as a function of current injection steps was also unaffected by FLG treatments (Fig. 5L).
FLG increases the response of DRG neurons to noxious stimuli
DRG neurons are polymodal, detecting and responding to distinct noxious stimuli through a variety of specific receptors.68 Since tissue acidification occurs under various painful conditions, primary sensory neurons express several acid sensors to detect changes in extracellular pH, such as acid-sensing ion channels (ASICs),69 TRPV1,70 proton-sensing G protein-coupled receptors, and two-pore domain potassium channels (K2P).71,72 Thus, we investigated whether exposure of DRG neurons to FLG alters their physiological responses to noxious stimuli. When the response of DRG neurons to mild acidic pH (pH 6.0) was investigated, FLG-treated DRG neurons displayed similar responses to CTRL neurons in terms of both activated inward current and membrane depolarization (Fig. S9A and B†). Additional experiments were conducted to verify the DRG response to mechanical stress with or without FLG treatment. Specifically, we tested neurons for the increase in membrane surface tension produced by osmotic swelling in terms of change in inward/outward currents. As previously seen in response to acidic pH, treating cells with hypotonic solution increases whole-cell current recorded at 80/−80 mV evoked by ramp stimulation. This effect is likely due to a general activation of several mechano-sensitive channels expressed in DRG neurons, such as TRPV1,73 TRPV4,74 TRPA1,75 TRPC5,76 TRPM8,77 TWIK-related spinal cord K+ channel (TRESK) of the K2P family78 and K+–Cl− cotransporter 3 (KCC3).79 Under these conditions, no significant difference was found in inward/outward currents in CTRL and FLG-treated neurons (Fig. S9C†).DRG neurons are also known to express several types of TRP channels that are implicated in a variety of sensory processes, such as thermosensation, mechanosensation, osmosensation, olfaction, taste, and pain perception.80,81 In particular, two key chemoreceptors identified in these neurons are the TRPV1, the receptor for “hot” compounds in different types of pepper,82–84 and TRPA1, the receptor for pungent compounds, such as mustard oil and cinnamon, that are expressed in small/medium neuronal subgroup.85,86 Consequently, we investigated whether the treatment with 10 μg mL−1 FLG alters the response of these two nociceptors in DRG neurons.
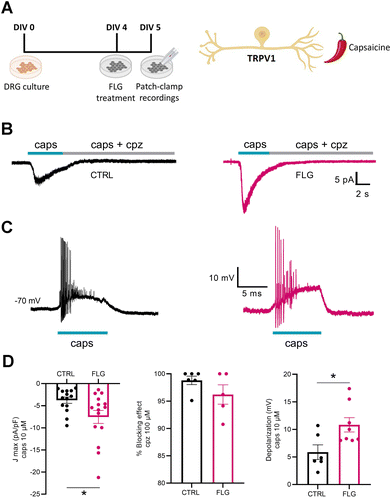
To investigate DRG responses mediated by TRPV1 channels, we used the agonist capsaicin (Fig. 6A). Interestingly, when we measured the capsaicin-dependent inward current at negative potentials (−70 mV) using whole-cell patch-clamp in voltage-clamp configuration, we observed a significantly increased response of neurons treated with FLG, compared with the control group (Fig. 6B–D). The specificity of the effect was assessed using capsazepine, a selective antagonist of the TRPV1 channel, that effectively blocked the inward current in both CTRL and FLG-treated neurons (Fig. 6D). In addition, current-clamp recordings showed that the observed increase in the capsaicin-evoked inward current at negative potentials was associated with a sustained depolarization of the membrane potential of FLG-treated DRG neurons (Fig. 6D).
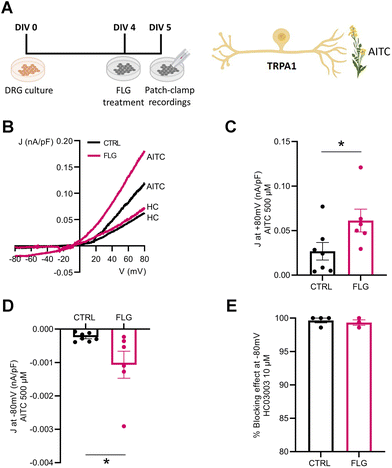
We also investigated the effect of FLG treatment on the responses of DRG neurons mediated by TRPA1 channels. To this purpose, we administered allyl isothiocyanate (AITC), a specific TRPA1 agonist, to DRG neurons in the presence and absence of FLG treatment (Fig. 7A). When the current density/voltage (J/V) relationships of the TRPA1-mediated current activated by AITC were studied in CTRL and FLG-treated DRG neurons using a ramp protocol (Fig. 7B), a significantly increased response was again observed in DRG-treated neurons compared to CTRL neurons, at both at +80 and −80 mV holding potentials (Fig. 7C and D). The specificity of the response for TRPA1 channels was assessed by using, under the same conditions, HC03003, a selective TRPA1 channel blocker, that fully abolished the AITC-induced current in both CTRL and FLG-treated neurons (Fig. 7E).
Taken together, all these results show that in the presence of both low and high FLG concentrations, the functionality of DRG neurons does not change in terms of acidic pH sensitivity, macroscopic currents, and firing properties. However, when we looked at the functionality of DRG neurons sensitive to specific irritant modalities, we found an increased response in DRG neurons treated with a low concentration of FLG.
As mentioned, TRP channels are highly expressed in a subset of small/medium diameter DRG neurons,80 the same subtypes analyzed in the electrophysiological experiments, and are a major integrator of noxious stimuli, ranging from exogenous chemical agents (capsaicin) to endogenous agonists (e.g., acidic and basic pH).87 To understand whether the observed effects were due to an increase of channel expression or to an increased responsivity of FLG-treated neurons to the stimulus, we looked at the TRPV1 expression by DRG neurons under our experimental conditions. When compared to control DRG neurons, FLG-treated neurons did not show a clear-cut overexpression of TRPV1/TRPA1 channels, nor a tendency of the channels to translocate from the membrane to the intracellular compartment (Fig. S10 and S11†). Proteomics data were also unable to uncover a clear upregulation of TRPV1 and TRPA1 channels. This can be due to the large heterogeneity of neuronal subtypes and the presence of glial cells that will dilute out alterations in the single-cell population on which electrophysiological recordings were performed. However, our data suggest that the increase of TRP-mediated currents in FLG-treated neurons is attributable to an increased membrane exposure of the cellular TRP pool or an enhancement of channel sensitivity to the agonists. The latter explanation is in line with the proteomics data of increased complement activation in FLG-treated neurons that might imply an increased sensitization of the nociceptive neurons (ESI file 3†). Additional research is needed to explain why FLG increases the activity of DRG neurons in response to certain physiological stimuli and to fathom the importance and the consequences of this effect.
Primary sensory neuron cultures were prepared from wild-type P2/P3 Sprague-Dawley rat pups (Charles River, Calco, Italy). The DRGs were dissected out, dissociated, and cultured with a protocol optimized in-house.89,90 We prepared two types of medium to be used during dissociation. The dissociation medium consisted of Neurobasal (Gibco, Thermo Fisher Scientific, Massachusetts, USA) supplemented with fetal bovine serum (10%, FBS, Thermo Fisher Scientific) and penicillin/streptomycin (50 U mL−1, Sigma-Aldrich, Missouri, USA). The growth medium consisted of Neurobasal supplemented with Pen/Strep (50 U mL−1), recombinant human β-NGF (50 ng mL−1, Bio-Techne, Minnesota, USA), BSA (0.05%), B-27 (2%, Thermo fisher scientific) and Glutamax (1%, Thermo Fisher Scientific).
After sacrificing the pups by decapitation, we made an incision from head to tail following the ribcage. We peeled the skin, cut out all organs, and trimmed the vertebrae to free up the spinal column. Once the spinal column was cut out, we used hydraulic extrusion to remove the spinal cord.90 We then cut through the ventral and dorsal columns to obtain two separate parts of the spinal column, each containing 15 DRGs. DRGs were now visible and ready to be extracted from their pockets. They were freed of excess nerve roots and meninges and washed in HBSS (Hank's Balanced Salt Solution, Sigma-Aldrich).
After the isolation of the DRGs, the dissociation could start. HBSS was removed and replaced by Trypsin–EDTA (0.05%, TE, Thermo Fisher Scientific), and DRGs were incubated for 20 min at 37 °C. Then, DRGs were collected in a 50 ml Falcon tube using a flamed Pasteur glass pipet and TE in the dissociation medium. DRGs were spun down at 200g for 2 min, and the supernatant was removed and replaced by collagenase (0.1%, Type 1, Sigma-Aldrich) for a 90 min dissociation at 37 °C. After this second dissociation step, collagenase was neutralized and removed by centrifugation (200g, 3 min). One mL of dissociation medium was added to the pellet, and DRGs were mechanically dissociated by triturating 6–10 times using a flamed glass Pasteur pipet. This was again centrifuged (200g, 5 min) and the dissociation medium was replaced by 1 mL of growth medium. DRGs were further mechanically dissociated until no DRGs were visible. Dissociated cells were spotted onto laminin/poly-D-lysine (PDL) (Mouse Laminin, Sigma; PDL, Sigma) coated glass coverslips, in 6- or 12-well plates (Falcon, Corning, NY, USA). After 1 h incubation to allow cells to attach, the well was flooded with the appropriate amount of medium. The final cell density was 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL. To reduce the presence of glial cells, the antimitotic cytosine β-D-arabinoside (final concentration of 1 μM) was added after 24 h and again at DIV4. This treatment was done by replacing half of the medium with fresh growth medium containing 2 μM cytosine β-D-arabinoside.
000 cells per mL. To reduce the presence of glial cells, the antimitotic cytosine β-D-arabinoside (final concentration of 1 μM) was added after 24 h and again at DIV4. This treatment was done by replacing half of the medium with fresh growth medium containing 2 μM cytosine β-D-arabinoside.
FLG treatment was initiated at DIV4 by replacing half of the medium with fresh growth medium containing either FLG at the desired concentration or H2O (CTRL). Neurons were exposed to graphene for a minimum of 4 h to a maximum of 7 days, at concentrations ranging from 10 to 100 μg mL−1. After exposure, DRG neurons were interrogated in terms of morphology, viability, proteomics, and electrophysiological properties.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 in 2% BSA. The primary antibodies used were rabbit polyclonal anti-β-tubulin III (Sigma Aldrich, T2200) and guinea pig polyclonal anti-GFAP (SYSY, 173004). Subsequently, neurons were washed 3 times in 1× PBS and incubated for 1 h at RT with secondary antibodies diluted 1
200 in 2% BSA. The primary antibodies used were rabbit polyclonal anti-β-tubulin III (Sigma Aldrich, T2200) and guinea pig polyclonal anti-GFAP (SYSY, 173004). Subsequently, neurons were washed 3 times in 1× PBS and incubated for 1 h at RT with secondary antibodies diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in BSA (2%). The fluorescently conjugated secondary antibodies were labeled by Alexa Fluor 488 or Alexa Fluor 568 (#11029 and #A11036, respectively; ThermoFisher Scientific, Molecular Probes). Finally, after another washing round, neurons were stained with Hoechst33342 diluted 1
500 in BSA (2%). The fluorescently conjugated secondary antibodies were labeled by Alexa Fluor 488 or Alexa Fluor 568 (#11029 and #A11036, respectively; ThermoFisher Scientific, Molecular Probes). Finally, after another washing round, neurons were stained with Hoechst33342 diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in BSA (2%) for 5 min.
100 in BSA (2%) for 5 min.
For studies on the channel distribution, polyclonal anti-TRPV1 (ACC-334, Alomone labs) and TRPA1 (ACC-037, Alomone labs) were used, and the permeabilization step with 0.1% Triton X-100 was not performed, in order to bind only to the extracellular domain. The stained coverslips were mounted in Vectashield (H-1000-10; Vector Laboratories, Newark, USA) on 1 mm thick microscope slides. Z-Stack image acquisition was performed using a confocal microscope (SP8, Leica Microsystems GmbH, Wetzlar, Germany) at 63× (1.4 NA) magnification.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g). The supernatants were collected, and the total protein content was determined with the BCA Protein Assay kit (Thermo Fisher Scientific) using the standard protocol.
000g). The supernatants were collected, and the total protein content was determined with the BCA Protein Assay kit (Thermo Fisher Scientific) using the standard protocol.
Total cell lysates were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C for 10 min. An aliquot of supernatant fractions was kept for the input (total) sample, while the remaining fraction was incubated with 100 μl of NeutrAvidin-conjugated agarose beads (Thermo Fisher Scientific) at 4 °C for 3 h. Supernatants containing intracellular proteins were collected, and the beads were washed 2× in RIPA buffer and 2× in PBS prior to samples (membrane proteins) elution.
000g at 4 °C for 10 min. An aliquot of supernatant fractions was kept for the input (total) sample, while the remaining fraction was incubated with 100 μl of NeutrAvidin-conjugated agarose beads (Thermo Fisher Scientific) at 4 °C for 3 h. Supernatants containing intracellular proteins were collected, and the beads were washed 2× in RIPA buffer and 2× in PBS prior to samples (membrane proteins) elution.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000). After washing with 0.05% TBS-T, the membrane was incubated with HRP-conjugated secondary antibodies (Abcam, 1
2000). After washing with 0.05% TBS-T, the membrane was incubated with HRP-conjugated secondary antibodies (Abcam, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) for 1 h at RT. The secondary antibodies were washed away in 0.05% TBS-T, and the membrane was developed using the ECL Prime Western Blotting System (Amersham Protran, Cytiva). The chemiluminescent signals were revealed using the iBright FL1500 Imaging System (Thermo Fisher Scientific, A44241), and band intensities were analyzed with iBright Analysis Software. Since TRPV1 and Na+/K+ ATPase α-1 have similar molecular weights, after the first acquisition, the nitrocellulose membranes were washed to remove the chemiluminescent substrate and incubated in Restore Western Blot Stripping Buffer (Thermo Fisher Scientific) for 20 min to remove primary and secondary antibodies, washed in 0.05% TBS-T and tested for removal of antibodies before performing the next immunoblot experiments.
5000) for 1 h at RT. The secondary antibodies were washed away in 0.05% TBS-T, and the membrane was developed using the ECL Prime Western Blotting System (Amersham Protran, Cytiva). The chemiluminescent signals were revealed using the iBright FL1500 Imaging System (Thermo Fisher Scientific, A44241), and band intensities were analyzed with iBright Analysis Software. Since TRPV1 and Na+/K+ ATPase α-1 have similar molecular weights, after the first acquisition, the nitrocellulose membranes were washed to remove the chemiluminescent substrate and incubated in Restore Western Blot Stripping Buffer (Thermo Fisher Scientific) for 20 min to remove primary and secondary antibodies, washed in 0.05% TBS-T and tested for removal of antibodies before performing the next immunoblot experiments.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolution, followed by MS/MS acquisition of 60 m/z transmission windows, each with a fixed 10 Da width. MS/MS spectra were acquired in HCD mode. The MS/MS spectra were analyzed using Spectronaut,91 by running a DirectDIA analysis against the reference Rattus FASTA database (Tax ID: 10
000 resolution, followed by MS/MS acquisition of 60 m/z transmission windows, each with a fixed 10 Da width. MS/MS spectra were acquired in HCD mode. The MS/MS spectra were analyzed using Spectronaut,91 by running a DirectDIA analysis against the reference Rattus FASTA database (Tax ID: 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 114 reporting 17
114 reporting 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 126 reviewed entries). Cysteine carbamidomethylation was selected as a fixed modification; acetylation, methylation, deamination, oxidation, and phosphorylation were selected as variable modifications. Positive protein identifications were retained at 1% false discovery rate (FDR) threshold and at least two peptides were used for protein quantification. Features with CV (coefficient of variation) > 30% in the QC samples were removed. The final data set was analyzed using the free on line software Metaboanalyst.92 Features with more than 50% of missing values were removed, data were normalized by sum, Log-transformed and Pareto scaled. Gene Ontology analysis was performed using STRING.93
126 reviewed entries). Cysteine carbamidomethylation was selected as a fixed modification; acetylation, methylation, deamination, oxidation, and phosphorylation were selected as variable modifications. Positive protein identifications were retained at 1% false discovery rate (FDR) threshold and at least two peptides were used for protein quantification. Features with CV (coefficient of variation) > 30% in the QC samples were removed. The final data set was analyzed using the free on line software Metaboanalyst.92 Features with more than 50% of missing values were removed, data were normalized by sum, Log-transformed and Pareto scaled. Gene Ontology analysis was performed using STRING.93
Conclusions
In conclusion, our data show excellent FLG biocompatibility towards DRG sensory neurons. Upon incubations with different FLG concentrations (10–100 μg mL−1), for various incubation times (1–7 days), neuronal viability, morphology, protein content, and basic electrical properties were preserved. Graphene flakes appear to be actively internalized and accumulated in large intracellular vesicles and possibly degraded over time.For the higher FLG concentration (100 μg mL−1), we observed mild morphological changes and protein dysregulation related to the glutamate signaling pathway and collagen. Glutamate is an essential neurotransmitter in the nervous system, and its upregulation can cause overexcitation of the neurons. Collagen also plays a pivotal role in peripheral nerves for Swann cell differentiation and nerve regeneration.88 This might be important when graphene is employed as a scaffold for nerve regeneration after injury. The complement pathway was also found to be upregulated, which in normal circumstances can lead to inflammatory damage, but in the case of nerve injury, it can stimulate nerve regeneration and sensitization of nociceptive neurons.95
Interestingly, in our electrophysiological investigation under noxious stimuli, we observed an increased functional response upon FLG treatment of the nociceptive subpopulation of DRG neurons in response to irritants specific for the TRPV1 and TRPA1 chemoreceptors involved in sensing acute noxious, mechanical, and chemical stimuli, as well as in neuropathic pain. Considering the present findings, the setup of future applications of FLG in regenerative medicine, tissue engineering, and drug delivery should consider the potential interference of this material with the physiology of these channels. Even when employed in planar substrates, if subjected to biological and mechanical stress, graphene can undergo aging and delamination over time. Therefore, the possibility should be considered that FLG flakes, in contact with sensory neurons, could lower their activation threshold (sensitization) and induce an increase in the perception of irritant stimuli. On the other hand, our work could potentially open up novel routes for applications of bare graphene materials. Further investigation of their specific role in sensory neurons’ overactivation might introduce the use of these materials under conditions in which nociception is impaired.
Author contributions
L. D., F. F., M. T., N. L.: investigation, formal analysis; L. D. writing; D. D.: investigation; E. V.: resources; S. V., F. L., A. A., V. C.: Formal analysis; P. V., V. C., F. B.: conceptualization, supervision, writing, review, and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Ilaria Dallorto, Rossana Ciancio, Diego Moruzzo, and Arta Mehilli, and Antonella Marte for administrative and technical help. The project has received funding from: the European Union's Horizon 2020 Research and Innovation Programme under Grant Agreement No. 881603 Graphene Flagship, Core 2 and Core 3 (to F. B.); the Italian Ministry of Foreign Affairs and International Cooperation (grant agreement no. MAE00694702021-05-20 to F. B.); IRCCS Ospedale Policlinico San Martino, Genova, Italy (Ricerca Corrente and “5 × 1000” to F. B. and V. C.).References
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Science, 2004, 306, 666–669 CrossRef CAS PubMed.
- K. S. Novoselov, D. Jiang, F. Schedin, T. J. Booth, V. V. Khotkevich, S. V. Morozov and A. K. Geim, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10451–10453 CrossRef CAS PubMed.
- A. K. Geim, Science, 2009, 324, 1530–1534 CrossRef CAS PubMed.
- Y. Zhang, T. R. Nayak, H. Hong and W. Cai, Nanoscale, 2012, 4, 3833–3842 RSC.
- J. Byun, J. Microbiol. Biotechnol., 2015, 25, 145–151 CrossRef CAS PubMed.
- D. Chimene, D. L. Alge and A. K. Gaharwar, Adv. Mater., 2015, 27, 7261–7284 CrossRef CAS PubMed.
- V. C. Sanchez, A. Jachak, R. H. Hurt and A. B. Kane, Chem. Res. Toxicol., 2012, 25, 15–34 Search PubMed.
- K. Kostarelos and K. S. Novoselov, Science, 2014, 344, 261–263 CrossRef CAS PubMed.
- M. Bramini, G. Alberini, E. Colombo, M. Chiacchiaretta, M. L. DiFrancesco, J. F. Maya-Vetencourt, L. Maragliano, F. Benfenati and F. Cesca, Front. Syst. Neurosci., 2018, 12, 12 CrossRef PubMed.
- J. W. Jeong, G. Shin, S. I. Park, K. J. Yu, L. Xu and J. A. Rogers, Neuron, 2015, 86, 175–186 CrossRef CAS PubMed.
- V. S. Polikov, P. A. Tresco and W. M. Reichert, J. Neurosci. Methods, 2005, 148, 1–18 CrossRef PubMed.
- M. P. Ward, P. Rajdev, C. Ellison and P. P. Irazoqui, Brain Res., 2009, 1282, 183–200 CrossRef CAS PubMed.
- K. Scholten and E. Meng, Lab Chip, 2015, 15, 4256–4272 RSC.
- A. Halim, K. Y. Qu, X. F. Zhang and N. P. Huang, ACS Biomater. Sci. Eng., 2021, 7, 3503–3529 CrossRef CAS PubMed.
- J. Chen, H. Liu, C. Zhao, G. Qin, G. Xi, T. Li, X. Wang and T. Chen, Biomaterials, 2014, 35, 4986–4995 CrossRef CAS PubMed.
- S. R. Shin, Y. C. Li, H. L. Jang, P. Khoshakhlagh, M. Akbari, A. Nasajpour, Y. S. Zhang, A. Tamayol and A. Khademhosseini, Adv. Drug Delivery Rev., 2016, 105, 255–274 CrossRef CAS PubMed.
- Z. Li, W. Zhang and F. Xing, Int. J. Mol. Sci., 2019, 20(10), 2461 CrossRef PubMed.
- H. Zhang, T. Fan, W. Chen, Y. Li and B. Wang, Bioact. Mater., 2020, 5, 1071–1086 Search PubMed.
- X. Wang, M. Guo, Y. Liu, K. Niu, X. Zheng, Y. Yang and P. Wang, ACS Appl. Bio Mater., 2021, 4, 4236–4243 CrossRef CAS PubMed.
- Y. Zheng, X. Hong, J. Wang, L. Feng, T. Fan, R. Guo and H. Zhang, Adv. Healthc. Mater., 2021, 10, e2001743 CrossRef PubMed.
- M. Aleemardani, P. Zare, A. Seifalian, Z. Bagher and A. M. Seifalian, Biomedicines, 2021, 10(1), 73 CrossRef PubMed.
- W. Guo, S. Wang, X. Yu, J. Qiu, J. Li, W. Tang, Z. Li, X. Mou, H. Liu and Z. Wang, Nanoscale, 2016, 8, 1897–1904 RSC.
- F. Emadi, A. Amini, A. Gholami and Y. Ghasemi, Sci. Rep., 2017, 7, 42258 CrossRef CAS PubMed.
- A. Rhazouani, H. Gamrani, M. El Achaby, K. Aziz, L. Gebrati, M. S. Uddin and F. Aziz, BioMed Res. Int., 2021, 2021, 5518999 Search PubMed.
- Z. Guo, P. Zhang, A. J. Chetwynd, H. Q. Xie, E. Valsami-Jones, B. Zhao and I. Lynch, Nanoscale, 2020, 12, 18600–18605 RSC.
- E. C. Qin, M. E. Kandel, E. Liamas, T. B. Shah, C. Kim, C. D. Kaufman, Z. J. Zhang, G. Popescu, M. U. Gillette, D. E. Leckband and H. Kong, Acta Biomater., 2019, 90, 412–423 CrossRef CAS PubMed.
- M. Lv, Y. Zhang, L. Liang, M. Wei, W. Hu, X. Li and Q. Huang, Nanoscale, 2012, 4, 3861–3866 RSC.
- P. Li, T. Xu, S. Wu, L. Lei and D. He, J. Appl. Toxicol., 2017, 37, 1140–1150 CrossRef CAS PubMed.
- M. Bramini, A. Rocchi, F. Benfenati and F. Cesca, Adv. Neurobiol., 2019, 22, 51–79 Search PubMed.
- M. Bramini, S. Sacchetti, A. Armirotti, A. Rocchi, E. Vazquez, V. Leon Castellanos, T. Bandiera, F. Cesca and F. Benfenati, ACS Nano, 2016, 10, 7154–7171 CrossRef CAS PubMed.
- M. Bramini, M. Chiacchiaretta, A. Armirotti, A. Rocchi, D. D. Kale, C. Martin, E. Vazquez, T. Bandiera, S. Ferroni, F. Cesca and F. Benfenati, Small, 2019, 15, e1900147 CrossRef PubMed.
- M. M. Ghazimoradi, F. Vahdatazad, F. Jalali and M. Rafieian-Kopaei, Curr. Pharm. Des., 2022, 28(44), 3572–3581 CrossRef CAS PubMed.
- K. Lawkowska, M. Pokrywczynska, K. Koper, L. A. Kluth, T. Drewa and J. Adamowicz, Int. J. Mol. Sci., 2021, 23(1), 33 CrossRef PubMed.
- G. Audira, J. S. Lee, P. Siregar, N. Malhotra, M. J. M. Rolden, J. C. Huang, K. H. Chen, H. S. Hsu, Y. Hsu, T. R. Ger and C. D. Hsiao, Environ. Pollut., 2021, 278, 116907 CrossRef CAS PubMed.
- E. G. Afshar, A. Zarrabi, A. Dehshahri, M. Ashrafizadeh, G. Dehghannoudeh, B. Behnam, A. Mandegary, A. Pardakhty, R. Mohammadinejad and S. Tavakol, FlatChem, 2020, 22, 100173 CrossRef.
- O. Akhavan, E. Ghaderi, E. Abouei, S. Hatamie and E. Ghasemi, Carbon, 2014, 66, 395–406 CrossRef CAS.
- M. Catanesi, G. Panella, E. Benedetti, G. Fioravanti, F. Perrozzi, L. Ottaviano, L. D. Leandro, M. Ardini, F. Giansanti, M. d'Angelo, V. Castelli, F. Angelucci, R. Ippoliti and A. Cimini, Nanomedicine, 2018, 13, 3091–3106 CrossRef CAS.
- A. Franceschi Biagioni, G. Cellot, E. Pati, N. Lozano, B. Ballesteros, R. Casani, N. C. Coimbra, K. Kostarelos and L. Ballerini, Biomaterials, 2021, 271, 120749 CrossRef CAS PubMed.
- M. G. Tupone, G. Panella, M. d'Angelo, V. Castelli, G. Caioni, M. Catanesi, E. Benedetti and A. Cimini, Int. J. Mol. Sci., 2021, 22(23), 13047 CrossRef CAS PubMed.
- G. Cellot, L. Jacquemin, G. Reina, A. Franceschi Biagioni, M. Fontanini, O. Chaloin, Y. Nishina, A. Bianco and L. Ballerini, ACS Appl. Nano Mater., 2023, 6, 8105 CrossRef CAS PubMed.
- A. H. Palejwala, J. S. Fridley, J. A. Mata, E. L. Samuel, T. G. Luerssen, L. Perlaky, T. A. Kent, J. M. Tour and A. Jea, Surg. Neurol. Int., 2016, 7, 75 CrossRef PubMed.
- T. A. Tabish, M. Z. I. Pranjol, H. Hayat, A. A. M. Rahat, T. M. Abdullah, J. L. Whatmore and S. Zhang, Nanotechnology, 2017, 28, 504001 CrossRef PubMed.
- Q. R. Zhang, X. L. Liu, H. Y. Meng, S. J. Liu and C. D. Zhang, Environ. Sci.: Nano, 2018, 5, 1361–1371 RSC.
- V. Castagnola, L. Deleye, A. Podesta, E. Jaho, F. Loiacono, D. Debellis, M. Trevisani, D. Z. Ciobanu, A. Armirotti, F. Pisani, E. Flahaut, E. Vazquez, M. Bramini, F. Cesca and F. Benfenati, Nano Lett., 2023, 23, 2981–2990 CrossRef CAS PubMed.
- A. A. John, A. P. Subramanian, M. V. Vellayappan, A. Balaji, H. Mohandas and S. K. Jaganathan, Int. J. Nanomed., 2015, 10, 4267–4277 CAS.
- R. Rauti, N. Lozano, V. Leon, D. Scaini, M. Musto, I. Rago, F. P. Ulloa Severino, A. Fabbro, L. Casalis, E. Vazquez, K. Kostarelos, M. Prato and L. Ballerini, ACS Nano, 2016, 10, 4459–4471 CrossRef CAS PubMed.
- Q. Liu, G. Liu, X. Liu, M. Yang, S. Xing, Y. Du and X. Xiong, Appl. Nanosci., 2020, 10, 687–694 CrossRef CAS.
- J. Wang, Y. Cheng, L. Chen, T. Zhu, K. Ye, C. Jia, H. Wang, M. Zhu, C. Fan and X. Mo, Acta Biomater., 2019, 84, 98–113 CrossRef CAS PubMed.
- C. Dong, F. Qiao, W. Hou, L. Yang and Y. Lv, Appl. Mater. Today, 2020, 21, 100870 CrossRef.
- Z. Huang, Z. Guo, M. Sun, S. Fang and H. Li, RSC Adv., 2019, 9, 28627–28635 RSC.
- D. Convertino, S. Luin, L. Marchetti and C. Coletti, Front. Neurosci., 2018, 12, 1 CrossRef PubMed.
- D. Convertino, F. Fabbri, N. Mishra, M. Mainardi, V. Cappello, G. Testa, S. Capsoni, L. Albertazzi, S. Luin, L. Marchetti and C. Coletti, Nano Lett., 2020, 20, 3633–3641 CrossRef CAS PubMed.
- T. Song, K. Gu, W. Wang, H. Wang, Y. Yang, L. Yang, P. Ma, X. Ma, J. Zhao, R. Yan, J. Guan, C. Wang, Y. Qi and J. Ya, J. Pharm. Sci., 2015, 104, 3851–3860 CrossRef CAS PubMed.
- Z. Zhang, X. Wang, P. Li, M. Bai and W. Qi, J. Biomater. Sci., Polym. Ed., 2021, 32, 874–885 CrossRef CAS PubMed.
- M. Hanani, Brain Res. Rev., 2005, 48, 457–476 CrossRef CAS PubMed.
- R. S. Nascimento, M. F. Santiago, S. A. Marques, S. Allodi and A. M. Martinez, Braz. J. Med. Biol. Res., 2008, 41, 1011–1017 CrossRef CAS PubMed.
- H. Liu, L. Zhao, W. Gu, Q. Liu, Z. Gao, X. Zhu, Z. Wu, H. He, F. Huang and W. Fan, J. Mol. Histol., 2018, 49, 257–263 CrossRef CAS PubMed.
- C. Braccia, V. Castagnola, E. Vazquez, V. J. Gonzalez, F. Loiacono, F. Benfenati and A. Armirotti, Carbon, 2021, 185, 591–598 CrossRef CAS.
- F. Alnasser, V. Castagnola, L. Boselli, M. Esquivel-Gaon, E. Efeoglu, J. McIntyre, H. J. Byrne and K. A. Dawson, Nano Lett., 2019, 19, 1260–1268 CrossRef CAS PubMed.
- M. Esquivel-Gaon, N. H. Nguyen, M. F. Sgroi, D. Pullini, F. Gili, D. Mangherini, A. I. Pruna, P. Rosicka, A. Sevcu and V. Castagnola, Nanoscale, 2018, 10, 6539–6548 RSC.
- M. Kucki, L. Diener, N. Bohmer, C. Hirsch, H. F. Krug, V. Palermo and P. Wick, J. Nanobiotechnol., 2017, 15, 46 CrossRef PubMed.
- H. Suzuki, T. Toyooka and Y. Ibuki, Environ. Sci. Technol., 2007, 41, 3018–3024 CrossRef CAS PubMed.
- S. Thongrattanasiri, F. H. Koppens and F. J. Garcia de Abajo, Phys. Rev. Lett., 2012, 108, 047401 CrossRef PubMed.
- O. Benzina, V. Szabo, O. Lucas, M. B. Saab, T. Cloitre, F. Scamps, C. Gergely and M. Martin, J. Biomed. Opt., 2013, 18, 106014 CrossRef PubMed.
- M. Martin, O. Benzina, V. Szabo, A. G. Vegh, O. Lucas, T. Cloitre, F. Scamps and C. Gergely, PLoS One, 2013, 8, e56286 CrossRef CAS PubMed.
- P. Valente, G. Lignani, L. Medrihan, F. Bosco, A. Contestabile, P. Lippiello, E. Ferrea, M. Schachner, F. Benfenati, S. Giovedi and P. Baldelli, J. Cell Sci., 2016, 129, 1878–1891 CAS.
- C. Prestigio, D. Ferrante, P. Valente, S. Casagrande, E. Albanesi, Y. Yanagawa, F. Benfenati and P. Baldelli, Mol. Neurobiol., 2019, 56, 6276–6292 CrossRef CAS PubMed.
- S. A. Scott, Sensory Neurons: Diversity, Development, and Plasticity, Oxford University Press, 1992 Search PubMed.
- R. Waldmann, G. Champigny, E. Lingueglia, J. R. De Weille, C. Heurteaux and M. Lazdunski, Ann. N. Y. Acad. Sci., 1999, 868, 67–76 CrossRef CAS PubMed.
- M. Petersen and R. H. LaMotte, Pain, 1993, 54, 37–42 CrossRef CAS PubMed.
- P. Holzer, Handb. Exp. Pharmacol., 2009, 194, 283–332 CAS.
- L. A. Pattison, G. Callejo and E. St John Smith, Philos. Trans. R. Soc. London, Ser. B, 2019, 374, 20190291 CrossRef CAS PubMed.
- T. Yamada, S. Ugawa, T. Ueda, Y. Ishida, K. Kajita and S. Shimada, J. Histochem. Cytochem., 2009, 57, 277–287 CrossRef CAS PubMed.
- N. Alessandri-Haber, J. J. Yeh, A. E. Boyd, C. A. Parada, X. Chen, D. B. Reichling and J. D. Levine, Neuron, 2003, 39, 497–511 CrossRef CAS PubMed.
- F. Fujita, K. Uchida, Y. Takayama, Y. Suzuki, M. Takaishi and M. Tominaga, J. Physiol. Sci., 2018, 68, 431–440 CrossRef CAS PubMed.
- A. Gomis, S. Soriano, C. Belmonte and F. Viana, J. Physiol., 2008, 586, 5633–5649 CrossRef CAS PubMed.
- T. Quallo, N. Vastani, E. Horridge, C. Gentry, A. Parra, S. Moss, F. Viana, C. Belmonte, D. A. Andersson and S. Bevan, Nat. Commun., 2015, 6, 7150 CrossRef PubMed.
- G. Callejo, J. P. Giblin and X. Gasull, PLoS One, 2013, 8, e64471 CrossRef CAS PubMed.
- B. Flores and E. Delpire, Cell. Physiol. Biochem., 2020, 54, 577 CrossRef CAS PubMed.
- C. Montell, Sci. STKE, 2005,(272), re3 Search PubMed.
- N. Damann, T. Voets and B. Nilius, Curr. Biol., 2008, 18, R880–R889 CrossRef CAS PubMed.
- M. J. Caterina, M. A. Schumacher, M. Tominaga, T. A. Rosen, J. D. Levine and D. Julius, Nature, 1997, 389, 816–824 CrossRef CAS PubMed.
- M. Tominaga and T. Tominaga, Pflugers Arch., 2005, 451, 143–150 CrossRef CAS PubMed.
- P. Valente, A. Fernandez-Carvajal, M. Camprubi-Robles, A. Gomis, S. Quirce, F. Viana, G. Fernandez-Ballester, J. M. Gonzalez-Ros, C. Belmonte, R. Planells-Cases and A. Ferrer-Montiel, FASEB J., 2011, 25, 1628–1640 CrossRef CAS PubMed.
- G. M. Story, A. M. Peier, A. J. Reeve, S. R. Eid, J. Mosbacher, T. R. Hricik, T. J. Earley, A. C. Hergarden, D. A. Andersson, S. W. Hwang, P. McIntyre, T. Jegla, S. Bevan and A. Patapoutian, Cell, 2003, 112, 819–829 CrossRef CAS PubMed.
- K. Nagata, A. Duggan, G. Kumar and J. Garcia-Anoveros, J. Neurosci., 2005, 25, 4052–4061 CrossRef CAS PubMed.
- M. J. Caterina, A. Leffler, A. B. Malmberg, W. Martin, J. Trafton, K. Petersen-Zeitz, M. Koltzenburg, A. Basbaum and D. Julius, science, 2000, 288, 306–313 CrossRef CAS.
- N. Liessi, L. Maragliano, V. Castagnola, M. Bramini, F. Benfenati and A. Armirotti, Anal. Chem., 2021, 93, 784–791 CrossRef CAS PubMed.
- D. E. Owen, Methods in Molecular Biology, in Neurotrophic Factors, 2012, 846(16), 179–187 CAS.
- M. Richner, S. B. Jager, P. Siupka and C. B. Vaegter, J. Visualized Exp., 2017, 22(119), 55226 Search PubMed.
- R. Bruderer, O. M. Bernhardt, T. Gandhi, S. M. Miladinović, L.-Y. Cheng, S. Messner, T. Ehrenberger, V. Zanotelli, Y. Butscheid and C. Escher, Mol. Cell. Proteomics, 2015, 14, 1400–1410 CrossRef CAS PubMed.
- Z. Pang, G. Zhou, J. Ewald, L. Chang, O. Hacariz, N. Basu and J. Xia, Nat. Protoc., 2022, 17, 1735–1761 CrossRef CAS PubMed.
- D. Szklarczyk, A. L. Gable, D. Lyon, A. Junge, S. Wyder, J. Huerta-Cepas, M. Simonovic, N. T. Doncheva, J. H. Morris and P. Bork, Nucleic Acids Res., 2019, 47, D607–D613 CrossRef CAS PubMed.
- A. E. Carpenter, T. R. Jones, M. R. Lamprecht, C. Clarke, I. H. Kang, O. Friman, D. A. Guertin, J. H. Chang, R. A. Lindquist, J. Moffat, P. Golland and D. M. Sabatini, Genome Biol., 2006, 7, R100 CrossRef PubMed.
- C. A. Warwick, A. L. Keyes, T. M. Woodruff and Y. M. Usachev, J. Biol. Chem., 2021, 297, 101085 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr03790h |
| ‡ Equal contribution. |
| § Co-last authors. |
| This journal is © The Royal Society of Chemistry 2024 |