Non-viral vector-based genome editing for cancer immunotherapy
Tianxu
Fang
ab and
Guojun
Chen
 *ab
*ab
aDepartment of Biomedical Engineering, McGill University, Montreal, QC H3G 0B1, Canada. E-mail: guojun.chen@mcgill.ca
bRosalind & Morris Goodman Cancer Institute, McGill University, Montreal, QC H3G 0B1, Canada
First published on 8th May 2024
Abstract
Despite the exciting promise of cancer immunotherapy in the clinic, immune checkpoint blockade therapy and T cell-based therapies are often associated with low response rates, intrinsic and adaptive immune resistance, and systemic side effects. CRISPR-Cas-based genome editing appears to be an effective strategy to overcome these unmet clinical needs. As a safer delivery platform for the CRISPR-Cas system, non-viral nanoformulations have been recently explored to target tumor cells and immune cells, aiming to improve cancer immunotherapy on a gene level. In this review, we summarized the efforts of non-viral vector-based CRISPR-Cas-mediated genome editing in tumor cells and immune cells for cancer immunotherapy. Their design rationale and specific applications were highlighted.
1. Introduction
Cancer immunotherapy is a promising strategy with better anti-tumor efficacy than the traditional chemo- and radiotherapies.1–4 The adaptive immune responses are to be boosted to remodel the tumor immunosuppressive microenvironment to inhibit tumor growth and defeat tumor recurrence.5–7 Antigen presenting cells (APCs), including dendritic cells and macrophages, recognize the tumor antigens and present the antigens to adaptive immune cells, like T cells, which recognize and kill the tumor cells.8–10 The delicate immune system relies on the expression and recognition of membrane proteins expressed on the tumor cells and immune cells, and the secreted cytokines and chemokines from these cells, which can all be the targets for the treatments to enhance the anti-tumor immunity by boosting the functioning of immune-active cells and inhibiting immunosuppressive cells.11–15Immune checkpoint blockade and T cell-based therapies are commonly used in the clinic now.16–21 Immune checkpoint inhibitors can block the negative immune regulators, such as programmed cell death 1 (PD-1) protein, programmed cell death ligand 1 (PD-L1), and cytotoxic T-lymphocyte associated protein 4 (CTLA-4), to reactivate the anti-tumor immune systems.22–25 T cell-based therapies, including tumor infiltrating lymphocyte therapy, transgenic T cell receptor-T cell therapy, and chimeric antigen receptor (CAR)-T cell therapy, function primarily by increasing the number of tumor-specific T cells against tumor cells.26–29 Although durable therapeutic efficacies of these therapies have been observed in a subset of cancer patients, the low response rate and the intrinsic and adaptive immune resistance in patients still remain.30–38 Thus, novel immunotherapeutic strategies are urgently needed.
Genome editing appears to be an effective strategy for cancer immunotherapy since it could induce long-term effects in the context of gene expression of immunoregulatory proteins, cytokines, and chemokines on/from the cells to remodel anti-cancer immunity, which could be beneficial over conventional immunotherapeutic strategies, such as immune checkpoint antibodies, plasmids, messenger RNA, or small interfering RNA and gene therapies for cancer immunotherapies.39–42 Compared with other conventional genome editing systems, such as zinc finger nucleases (ZFNs) and transcription activator-like nucleases (TALEN), the CRISPR-Cas-based approaches, an emerging technology which won the 2020 Nobel Prize in Chemistry, show unique advantages.39,43–46 The CRISPR-Cas system, composed of Cas nuclease and guide RNA (gRNA),47–49 can easily modify targeted genes by designing gRNAs.50–54 The CRISPR-Cas technology also offers increased precision and efficiency with a reduced risk of off-target editing.39,55–58 Additionally, various new CRISPR-Cas systems, such as prime-editing and base-editing, create new opportunities for precise gene corrections.59–62
The implementation of CRISPR-Cas technology can take the form of plasmid encoding both Cas and sgRNA, CRISPR mRNA/sgRNA, or ribonucleoprotein (RNP, the complex of Cas protein and sgRNA).63–69 The plasmid DNA, RNA, and RNP all exhibit negative surface charges and low stability, leading to no to extremely low gene editing efficiency.70–75 Therefore, the development of nano-systems for the delivery of CRISPR-Cas systems is urgently needed to elevate the intracellular delivery efficiency and broaden the applications of CRISPR-Cas technology.
Viral vectors, such as adeno-associated viruses (AAVs) and lentiviral vectors (LVs), have been commonly employed to load and transport genome editing systems in cells efficiently.76–80 However, viral vectors are associated with many drawbacks, including immunogenicity, insertional mutagenesis, toxicity, and packing limitations.81–88 On the other hand, non-viral vectors like nanoparticles (NPs) are considered and investigated to overcome those limitations. Several unique NPs have been reported for CRISPR-Cas delivery, including polymeric, lipid, gold, and silicon nanoparticles.75,86,89–95 These NPs exhibit chemical design flexibility, high loading capacity, high safety and stability, biocompatibility, and low immunogenicity.71,94,96,97 In addition, NPs can be easily modified to target specific cells, making genome editing possible in a broad spectrum of cells in the complicated tumor microenvironment (TME) and immune systems.94,98–100
In this review, we reviewed the development of non-viral nanoformulations for the CRISPR-Cas system delivery in tumor cells and immune cells and highlighted their rationales and applications in cancer immunotherapy (Fig. 1). In tumor cells, gene editing targeted to the classical immune checkpoint, PD-L1, for enhanced T cell recognition and killing effect, and to other immune-related proteins for the promoted phagocytosis of tumor cells by macrophages and the preservation of vitality of T cells. As for gene editing in immune cells, we introduced the applications to macrophages, dendritic cells, and T cells. The goals of targeting macrophages lay in the induction of macrophage polarization from M2 to M1 phenotype, and the promotion of macrophage phagocytosis. The gene editing in dendritic cells enhanced the presentation of antigens to T cells, and in T cells strengthened the recognition of tumor cells for higher killing efficiency.
2. Genome editing targeting tumor cells
Tumors can escape immune surveillance through various mechanisms, in which the expression of immune checkpoints and the secretion of some immunomodulatory molecules have been widely investigated. For example, immune checkpoints which are expressed on tumor cell surfaces can interact with immune checkpoint receptors on immune cells to inhibit cell killing.101–103 Tumor cells also secrete immunosuppressive molecules to inhibit the activation and infiltration of immunosupportive cells (e.g., CD4+ and CD8+ T cells) and promote immunosuppressive cells (e.g., regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs)).104–109 Knockout of immune checkpoint expression and immunosuppressive molecule secretion in tumors using CRISPR-Cas tools could favor anti-tumor immunity (Table 1).| Target gene locus | CRISPR-Cas9 format | NP composition | Applications | Administration route | Ref. |
|---|---|---|---|---|---|
| PD-L1 | RNP | Ni2+-modified mesoporous PDA-covering Fe3O4 NPs | In vivo (B16F10 melanoma in C57BL/6 mice) | Intravenous injection | 123 |
| PD-L1 | RNP | Mesoporous silica NP-based virus-like NPs | In vivo (B16F10 melanoma in C57BL/6 mice) | Intravenous injection | 124 |
| PD-L1; MHC-1 | Plasmid | Polyplex of CRISPR plasmid DNA (pDNA) and thioketal-cross-linked polyethylenimine derivative, covered with glucose-modified poly(ethylene glycol)-b-polylysine | In vivo (B16F10 melanoma in C57BL/6 mice; B16F10 or 4T1 TNBC in BALB/c mice) | Intravenous injection | 125 |
| PD-L1 | Plasmid | Au nanoparticle-loaded core–shell tecto-dendrimers (Au CSTDs), gold NPs entrapped in lactobionic acid (LA)-modified G5 PAMAM dendrimers, covered with PBA-conjugated G3 PAMAM dendrimers, complexed with Cas9-PD-L1 pDNA | In vivo (B16F10 melanoma in C57BL/6 mice) | Intratumoral injection | 129 |
| PD-L1; PVR | RNP | Sequence-defined oligo(ethylenamino) amide-based carriers modified with folic acid | In vivo (CT26 colorectal carcinoma in BALB/c mice) | Intratumoral injection | 130 |
| PD-L1 | Plasmid | Polyplex of CRISPR/Cas13a pDNA and 4-(hydroxymethyl)phenylboronic acid (HPBA)-modified PEI, covered with cis-aconitic anhydride and sodium glucoheptonate dehydrate-modified PEG-b-polylysine | In vivo (B16F10 melanoma in C57BL/6 mice) | Intravenous injection | 137 |
| PD-L1 | mRNA | LNP | In vivo (ID8-luc ovarian cancer in C57BL/6 mice; MYC-driven liver cancer in transgenic mice) | Local injection, intravenous injection | 142 |
| CD47 | Plasmid | PEI-decorated gold nanoparticles | In vivo (B16F10 melanoma in C57BL/6 mice) | Peritumoral injection | 154 |
| CD47 | Plasmid | Nanocomplex of plasmids and fluorinated polyethylenimine, coated with hyaluronic acid and TME-sensitive peptides | In vivo (B16F10 melanoma in C57BL/6 mice) | Intravenous injection | 155 |
| CD47 | RNP | Cas9 RNP coated by N-(3-aminopropyl) methacrylamide (cationic monomer), methacrylate (anionic monomer), acrylated pheophorbide a (ICD monomer), acrylated PEG (APEG, blood-circulated monomer), folic acid modified APEG (tumor-targeted monomer), and N,N′-bis(acryloyl) cystamine (GSH-degradable crosslinker) | In vivo (B16F10 melanoma in C57BL/6 mice) | Intravenous injection | 159 |
| Ptpn2 | Plasmid | Nanocomplex of plasmids, branched PEI, and iRGD peptides, assembled with 3-(carboxypropyl) triphenyl phosphonium bromide-PEI-Ce6 | In vivo (B16F10 melanoma in C57BL/6 mice) | Intravenous injection | 168 |
| Ptpn2 | Plasmid | DBCO decorated liposome | In vivo (B16F10 melanoma in C57BL/6 mice) | Intravenous injection | 169 |
| SLC43A2 | Plasmid | Mn/Zn-ZIF-8 MOF | In vivo (4T1 TNBC in BALB/c mice) | Intravenous injection | 172 |
| HSP70; BAG3 | Plasmid | LEGEND composed of poly(β-amino ester) PAE-C14, semiconducting polymer BDT-TQE, tumor-targeting PEGylated lipids DSPE-PEG-AEAA; FUGEND composed of PDA subunit, poly(β-amino ester) PAE-C14, PEGylated lipids DSPE-PEG-AEAA | In vivo (B16F10 melanoma, LLC lung cancer, Hepa1-6 liver cancer in C57BL/6 mice; A549 lung cancer cells in Nod/SCID/IL2RG−/− mice; PDX of liver cancer in B-NDG B2m/KO mice plus (B2m(v2) (mut/mut), B2m (−/−) IL-2rg (−/−))) | Intravenous injection | 183 |
2.1. Targeting PD-L1
PD-L1 is one of the immune checkpoints that is often overexpressed on tumor cells, such as gastric cancer, ovarian cancer, melanoma, breast cancer, and colorectal cancer.110–117 Binding PD-L1 and programmed cell death-1 (PD-1) on T cells will lead to impeding of the T cell-mediated anti-immune responses.22,118–122 To this end, Lu et al.123 utilized Ni2+-modified mesoporous polydopamine (mPDA)-covering Fe3O4 NPs (termed FPP) to deliver Cas9 RNP targeting the PD-L1 gene (Fig. 2A). The magnetic targeting effect of the Fe3O4 NPs improved the accumulation of Cas9 RNP-loaded nanoparticles in tumor sites. Afterwards, near-infrared (NIR) light initiated the release of Cas9 RNP by disrupting the non-covalent bond between the metal ions and PDA. In B16F10 cells, FPP/RNP with NIR induced 42.1% of PD-L1 gene knockout. In the melanoma model in vivo, the gene knockout efficiency of FPP/RNP with NIR was about 25.1%, which translated to the best control of the tumor growth in this group. The treatment of FPP/RNP with NIR laser irradiation effectively reversed the immunosuppression of the TME and induced strong anti-tumor immune responses in vivo, including DC maturation, T cell activation and secretion of antibodies and cytokines. | ||
| Fig. 2 Editing of the PD-L1 gene in tumor cells for cancer immunotherapies. (A) FPP preparation and RNP loading, and illustration of the multi-functional photothermal therapy and CRISPR/Cas9 genome-editing strategy for editing PD-L1 gene and remodeling the immunosuppressive tumor environment in vivo.123 (B) Schematic illustration for the synthesis of Cas9-sgPD-L1-loaded VLN and the delivery process after intravenous injection, the suppression of Tregs and the activation of CD8+ T cells by the Cas9-sgPD-L1-loaded VLN for efficient cancer immunotherapy.124 | ||
Liu et al.124 presented a mesoporous silica nanoparticle (MSN)-based virus-like nanoparticle (denoted as VLN) as a multifaceted nanoplatform to co-deliver the CRISPR/Cas9 system and small molecule drugs (Fig. 2B). The RNP was conjugated on the surface-thiolated MSN via glutathione (GSH)-responsive disulfide bonds. The CRISPR/Cas9 system with an sgRNA targeting the PD-L1 encoding gene (denoted as sgPD-L1) was released in cytosol with high GSH present. VLN-sgPD-L1 decreased PD-L1 expression to 41.3% in vitro and exhibited 45.1% of PD-L1 knockout efficiency in vivo in B16F10 tumors. VLN-sgPD-L1 treatment significantly suppressed tumor growth and increased survival, as well as revitalized the exhausted T cells for enhanced anti-tumor immune activation.
Xing et al.125 developed a pH/photo dual-activatable binary CRISPR nanomedicine (denoted as DBCN), where the polyplex of CRISPR plasmid DNA (pDNA that targets PD-L1) and thioketal (TK)-cross-linked polyethylenimine derivative (modified with phenylboronic acid (PBA) and photosensitizer pheophorbide a (Pha), TK-PPP) formed the core and glucose-modified poly(ethylene glycol)-b-polylysine (mPEG113-b-PLys25/Glu) formed the shell (Fig. 3A). The core was exposed because of the disassembly of the shell in the acidic TME. PBA groups on the TK-PPP/pDNA polyplex can then bind with sialic acid (SA) overexpressed on the tumor cells to enhance tumor accumulation and cellular internalization of the CRISPR components.126 With tumor progression, MHC-1 expression on the tumor cell surface gradually decreases, resulting in insufficient tumor-associated antigen (TAA) presentation and immune evasion.127,128 Thus, the up-regulation of MHC-1 expression is supposed to effectively restore TAA presentation on the surface of tumor cells. The pDNA respectively encoding the CRISPR-Cas9 system for disrupting PD-L1 and the aCRISPR-dCas9 system for up-regulating MHC-1 was co-loaded and co-delivered into B16F10-OVA cells. Both PD-L1 disruption and MHC-1 up-regulation were observed after treatment with DBCN under pH 6.5 with laser irradiation, initiating robust T cell-dependent antitumor immune responses to inhibit the growth of both primary and distant tumors. In B16F10 tumor-bearing mice, the DBCN (with laser) treatment significantly controlled the growth of primary tumors and prolonged the survival of tumor-bearing mice. Immunological analysis showed that DBCN (with laser) treatment promoted the intratumoral infiltration of CD8+ T cells from 8.57% to 29.4% compared with PBS treatment. In addition, DBCN further triggered lasting and specific antitumor immune memory effects, which significantly prevented the recurrence and metastasis of malignant tumors. DBCN (with laser) treatment increased the fraction of effector memory T cells from 12.5% to 28.7% and central memory T cells from 18.7% to 32.8% compared with PBS treatment.
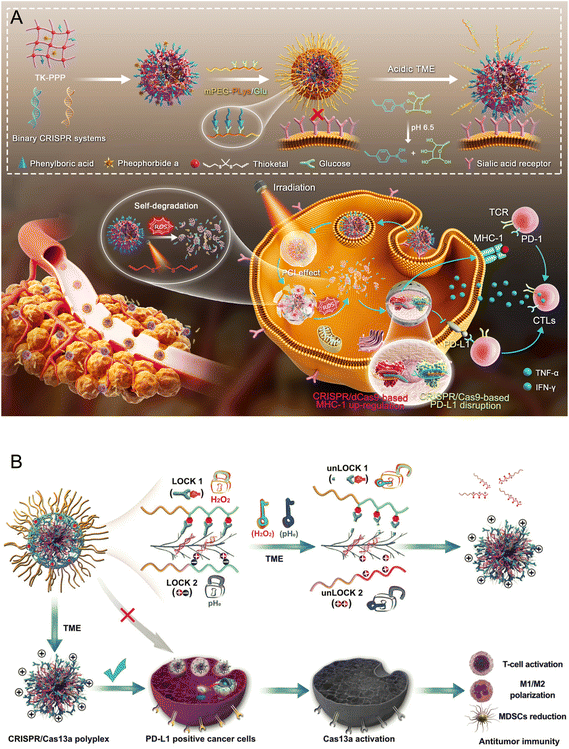 | ||
| Fig. 3 Editing of the PD-L1 gene in tumor cells for cancer immunotherapies. (A) Preparation of DBCN, and the schematic illustration of DBCN delivering binary CRISPR systems to simultaneously correct the disturbance of MHC-1 and PD-L1 expression to remodel tumor immunogenicity for efficient cancer immunotherapy.125 (B) Schematic illustration of DLNP for effective cancer immunotherapy. The CRISPR/Cas13a system can only be released from DLNP in a microenvironment with both a low pHe and high H2O2 concentration.137 | ||
Liu et al.129 constructed TME-responsive Au nanoparticle-loaded core–shell tecto dendrimers (Au CSTDs), composed of lactobionic acid (LA)-modified G5 poly(amidoamine) (PAMAM) dendrimer-entrapping Au NPs as the core and PBA-conjugated G3 PAMAM dendrimers assembled as the shell (i.e., the vector), to electrostatically complex and deliver Cas9-PD-L1 pDNA (Cas9-PD-L1). In B16F10 cells, the vector/Cas9-PD-L1 group showed the lowest expression level of PD-L1 proteins. In the melanoma mouse model, the vector/Cas9-PD-L1 group displayed a significantly better suppression of tumor growth and more significant PL-L1 expression inhibition than PD-L1 antibody treatment. Vector/Cas9-PD-L1 treatment also led to the increased distribution of CD4+/CD8+ T cells, decreased immunosuppressive cell populations, and increased levels of tumor necrosis factor α (TNF-α), interferon γ (IFN-γ) and interleukin (IL)-6 cytokines.
Lin et al.130 loaded Cas9/sgPD-L1 RNP and Cas9/sgPVR RNP in sequence-defined oligo(ethylenamino)amides (OAAs)-based carriers with folic acid (FolA) modification on the surface. The FolA mediated the Cas9 RNP delivery into tumor cells expressing folate receptor α (FRα).131 The T cell immunoreceptor with immunoglobulin (Ig) and immunoreceptor tyrosine-based inhibitory motif domain (TIGIT)/poliovirus receptor (PVR) is a newly identified immune checkpoint axis that emerged as a promising immunological target.132–134 Blocking the TIGIT/PVR axis has been proved to enable the reversal of T cell exhaustion and enhance antitumor efficacy in diverse types of cancer.132,135,136 A 54.4% dual knockout of PD-L1 and PVR in vitro and around 25% of dual knockout in vivo were detected in CT26 cells after treatment with FolA-PEG-nanocarriers. FolA-PEG-nanocarriers exhibited delayed tumor growth and enhanced CD8+ T cell infiltration in tumors.
Zhang et al.137 also established a tumor-targeting dual-locking nanoparticle (DLNP) to deliver the CRISPR/Cas13a system (Fig. 3B). Cas13a, also as a nuclease, targets RNA instead of DNA (which is targeted by Cas9), realizing temporary but safe gene regulation.138,139 The solid TME features a lower pH and higher ROS level, to which the DLNP was responsive to degrade into cationic polymers, facilitating the cellular internalization of the CRISPR/Cas13a system and gene editing activation.140,141 The crRNA targeting the PD-L1 gene was complexed in the CRISPR/Cas13a system. In the B16F10 melanoma mouse model, the DLNP group showed a superior control of tumor growth, where 50% of mice survived to 36 days. DLNP also induced 6-fold higher CD8+ T cell infiltration, 1.81-fold higher IFN-γ, and 3.84-fold higher TNF-α levels compared with the untreated group.
Zhang et al.142 reported a lipid nanoparticle (LNP) to deliver focal adhesion kinase (FAK) siRNA (siFAK), Cas9 mRNA, and PD-L1 sgRNA. FAK is frequently overexpressed in tumors, and promotes the adhesion of tumor cells to the tumor stroma and the ECM.143 The inhibition of FAK led to a decreased stiffness of the tumor ECM and enhanced infiltration of CD8+ cytotoxic T cells.144,145 In mice bearing ID8-luc xenograft ovarian tumors, siFAK + CRISPR-PD-L1-LNPs enhanced the infiltration of immune cells, especially T cells, which translated to the best anti-tumor effect. Similarly, the prolonged survival of mice and boosted infiltration of T cells and macrophages were discovered in the mouse model of liver cancer after siFAK + CRISPR-PD-L1-LNP treatment.
2.2. Targeting CD47
CD47 is a novel macrophage immune checkpoint expressed on cancer cells.146–148 CD47 can react with signal regulatory protein-α (SIRP-α) on macrophages to help cancer cells to bypass phagocytosis.149–153 Huang et al.154 utilized PEI-decorated gold nanoparticles to load the CRISPR/Cas9 plasmid pH330/sgCD47, obtaining a nanosystem termed AuPpH330/sgCD47. CD47 genomic disruption by the CRISPR/Cas9 plasmid was driven by a heat shock protein 70 (HSP70) promoter under NIR-II light. In in vitro experiments, the expression of CD47 protein on the surface of B16F10 cells was significantly downregulated in the AuPpH330/sgCD47 (with NIR irradiation) group. AuPpH330/sgCD47 (with NIR irradiation) triggered a powerful anticancer immune response, including M2-to-M1 phenotype macrophage polarization, CD8+ T cell infiltration, and Treg consumption. An immune memory response was also elicited, with the increased ratio of effector memory T cells and central memory T cells in the spleen.Lin et al.155 prepared an environment-responsive gene delivery system (abbreviated as HPT-PFs) for CD47 blockade with IL-12 production in tumor cells (Fig. 4A). The plasmids encoding IL-12 and Cas9/sgRNA were complexed with fluorinated polyethylenimine to form the core. The core was further coated by hyaluronic acid (HA) together with TME-sensitive peptides (TMSP, composed of cell-penetrating peptides (CPP) and shielding peptides connected by a matrix metalloproteinases-2/9 (MMP-2/9)-cleavable peptide linker (PVGLIG)). MMPs are overexpressed in tumors and can cleave the PVGLIG linkers to expose the CPP.156 The hyaluronic acid can interact with CD44 overexpressed on multiple tumor cells for active tumor targeting.157,158 HPT-PFs treatment exhibited the effective control of tumor growth and decreased CD47 expression in a melanoma mouse model. The anti-tumor effect was further enhanced by the co-delivery of IL-12 plasmid, which promoted the polarization of macrophages from M2 to M1 phenotype.
 | ||
| Fig. 4 Editing of the CD47 gene in tumor cells for cancer immunotherapies. (A) Schematic demonstration of HPT-PFs preparation and the nanoparticles’ endosome escape, IL-12 production, Cas9/sgRNA complex expression, and CD47 gene knockout in cells. CD47 blockade combined with IL-12 production synergistically promoted the polarization of TAMs for eliciting macrophage-mediated immunotherapy.155 (B) Schematic manifestation for Cas9NC fabrication and its degradation responding to GSH, and the delivery process of Cas9NC in cells and a simplified mechanism of Cas9NC-mediated antitumor immunity activation to inhibit the growth of both primary and abscopal tumors, preventing tumor recurrence and distant metastasis.159 | ||
Xing et al.159 prepared a Cas9 RNP nanocapsule (Cas9NC) formed by coating CD47-disrupting Cas9 RNP with acrylated pheophorbide a (APPa, the photosensitizer for photodynamic therapy) and multiple functional and crosslinked monomers, which endowed the Cas9NC with excellent blood stability, tumor targeting, laser-triggered immunogenic cell death (ICD), GSH-responsive degradability, and protection from enzyme degradation (Fig. 4B). Cas9NC (with laser) treatment of B16F10 cells resulted in 62.2% CD47 disruption. Cas9NC intravenous injections into melanoma-bearing mice led to a significant decrease in CD47 expression, which can promote the phagocytosis of tumor cells by dendritic cells and facilitate dendritic cell maturation and CD8+ T cell activation. The anti-tumor immune activation benefited the anti-metastatic efficiency of the Cas9NC treatments.
2.3. Targeting Ptpn2
Protein tyrosine phosphatase non-receptor type2 (Ptpn2) is a phosphatase involved in various signaling processes and it is frequently mutated in multiple malignancies.160–163 Deletion of Ptpn2 in tumor cells could elevate the immunotherapeutic efficacy by increasing the activated cytotoxic CD8+ T cells and IFN-γ mediated effects.164–167 Yang et al.168 prepared a positively charged “core” (termed PR@CCP) co-loaded with a modified mitochondria-targeting chlorin e6 (TPP-PEI-Ce6) and the CRISPR-Cas9 system targeting the Ptpn2 gene (Cas9-Ptpn2) (Fig. 5A). The PR@CCP core was further coated with a negatively charged HA “shell” by electrostatic adsorption to obtain the nanosystem, named HPR@CCP. The HPR@Cas9-Ptpn2 significantly inhibited Ptpn2 expression and elevated the percentages of CD3+CD8+ T cells in tumor tissues (∼35.29% compared with ∼25.25% in the PBS group) and blood (∼32.60% compared with ∼26.57% in the PBS group). | ||
| Fig. 5 Editing of the Ptpn2 gene in tumor cells for cancer immunotherapies. (A) The construction of HPR@CCP nanoparticles and their versatile application in cancer combined therapy.168 (B) Process of the selective labeling of tumor tissues via metabolic engineering by Ac4ManNAz/H-MnO2@Gel, and the illustration of the targeted CRISPR/Cas9 system delivery and tumor-specific gene editing of Ptpn2 by DBCO-Lipo/p via biorthogonal click chemistry.169 | ||
Yang et al.169 utilized the biorthogonal reaction to form a tumor-targeted CRISPR/Cas9 system delivery (Fig. 5B). Specifically, they developed an intelligent biodegradable hollow manganese dioxide (H-MnO2) nanoplatform for tumor-specific labeling by N-azidoacetylmannosamine-tetraacylated (Ac4ManNAz) through metabolic engineering. Then dibenzocyclooctyne (DBCO)-decorated liposome loading CRISPR/Cas9 plasmids (DBCO-Lipo/p) were used to disrupt the Ptpn2 gene in tumor cells through in vivo click chemistry. The combined treatment of Ac4ManNAz/H-MnO2Gel + DBCO-Lipo/p to B16F10 cells led to 28.2% of Ptpn2 gene disruption. In a melanoma mouse model, the Ac4ManNAz/H-MnO2Gel@DBCO-Lipo/p treatment group exhibited a gene editing efficiency of ∼25% and attenuated tumor growth of 75%. Moreover, significantly elevated tumor-infiltrating CD8+ T cells, transforming growth factor β (TGF-β), and IL-10 were observed, demonstrating that a robust anti-tumor immune response was effectively triggered.
2.4. Targeting SLC43A2
Methionine is a crucial protein for CD8+ T cells because methionine deficiency can induce CD8+ T cell death and dysfunction.170 However, the acquisition of sufficient methionine by CD8+ T cells can be interfered with by competitive methionine uptake in tumor cells through the high-expressing methionine transporter SLC43A2.171 Therefore, downregulation of SLC43A2 expression on tumor cells could promote the survival and activity of CD8+ T cells. Huang et al.172 encapsulated the CRISPR/Cas9 plasmid targeting SLC43A2 into Mn/Zn bimetallic metal–organic framework (MOF) nanoparticles to obtain PMZH (Fig. 6A). PMZH treatment was able to disturb the expression of SLC43A2 in 4T1 cells in vitro, which led to improved survival of cytotoxic T cells in the coculture system. PMZH exhibited 85.47% of 4T1 breast cancer growth inhibition in vivo, and significantly enhanced CD8+ T cell infiltration in tumors. | ||
| Fig. 6 (A) Editing of the SLC43A2 gene in tumor cells for cancer immunotherapies. Schematic illustration of PMZH nanoplatform formation and PMZH for methionine metabolism regulation, nutritional metal ion therapy, and immune stimulation.172 (B) Editing of the HSP70 gene in tumor cells for cancer immunotherapies. Schematic demonstration of the synthesis of LEGEND or FUGEND, and LEGEND- or FUGEND-mediated intracellular genome editing of tumor cells mediated by LEGEND or FUGEND and TME modulation to promote the adoptive T cell efficacy synergistically.183 | ||
2.5. Targeting to HSP70
HSP70 and Bcl-2-associated athanogene 3 (BAG3) are highly expressed in various cancer types and their intracellular activities have a great impact on the survival of tumor cells.173–175 HSP70 correlates with chemotherapy resistance, metastasis, tumor grade, and poor prognosis, while BAG3 is implicated in HSP70-related cell signaling.176 The overexpression of HSP70 and its co-chaperone BAG3 exhibits an anti-apoptotic mechanism in tumor cells so that these tumor cells can protect themselves from various stresses (such as chemotherapy and hypoxia) and T-cell-mediated killing effects.177–179 Besides, HSP70 and BAG3 also have the capacity to dynamically influence the TME, thereby fostering the advancement of cancer and treatment resistance, including T cell-based adoptive cell transfer.180–182To this end, Chen et al.183 established a nanoformulation that can respond to near-infrared (NIR) light or focused ultrasound (FUS) to deliver the heat-inducible CRISPR-Cas9 system that targets the HSP70 and BAG3 genes in tumor cells (Fig. 6B). The light-enabled genome-editing nanodevice (LEGEND) included poly[[diisopropyl 4-(4,8-bis(heptan-3-yloxy) benzo[1,2-b:4,5-b′]dithiophen-2-yl)]-alt-co-[[1,2,5]thiadiazolo[3,4-g]quinoxaline-6,7-dicarboxylate]], a semiconducting polymer which can convert NIR light-to-heat, and a heat-triggered Cas9 plasmid that encoded both HSP70 sgRNA and BAG3 sgRNA driven by a heat-shock promoter. Thus, NIR-II can induce the expression of plasmid loaded in LEGEND. Compared with the non-pretreated B16F10 tumors, the LEGEND pretreatment led to the increasing of infiltrating tumor-infiltrating lymphocytes (TILs) by 85 fold, and a higher proliferation rate (97.6%) than that in the non-pretreated tumors (13.6%). The notable anti-tumor therapeutic efficacy of LEGEND-sensitized adoptive TIL (LEGEND + TIL) was seen in B16F10 melanoma, and mouse models xenografted with A549 lung tumor and Hepa1-6 hepatocellular carcinoma cells. LEGEND pretreatment also led to more CAR-T cell infiltration in A549 primary and distant tumors, showing effective tumor growth control. In the humanized patient-derived xenograft (PDX) mouse model, LEGEND-sensitized adoptive CAR-T-cell therapy greatly enhanced the CAR-T cell infiltration into the PDX tumor, which was 6.5-fold higher than the group with CAR-T. FUS-enabled genome-editing nanodevice (FUGEND) included the sonosensitive 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-polyethylene glycol (DSPE-PEG). FUS can precisely generate hyperthermia in local deep tissues that light cannot reach. Hence, FUGEND can be applied to treat deep orthotopic tumors. Under ultrasound, the FUGEND system succeeded in promoting TIL proliferation and infiltration in orthotopic hepatocellular carcinoma.3. Genome editing targeting innate immune cells
Innate immune cells include macrophages, dendritic cells, neutrophils, and NK cells.184,185 Genome editing in these innate immune cells has potential to induce activation, polarization, or other immune activation-oriented transformation for an enhanced effect of killing tumor cells. In this section, we will introduce CRISPR system delivery via non-viral nanocarriers for genome editing in innate immune cells (mainly macrophages) for cancer immunotherapy (Table 2).| Target gene locus | Target immune cell | CRISPR-Cas9 format | NP composition | Applications | Administration route | Ref. |
|---|---|---|---|---|---|---|
| PI3Kγ | Macrophages | RNP | Nanovesicles derived from E. coli protoplasts and functionalized with a pH-responsive PEG-conjugated phospholipid derivative of DSPE-hydrazone bond-PEG2000 and a TAM-targeted phospholipid derivative of DSPE-galactosamine | In vivo (4T1 TNBC in BALB/c mice; MC38 colorectal cancer in C57BL/6J mice) | Intravenous injection | 195 |
| PI3Kγ | Macrophages | Plasmid | Exosomes externally engineered with a macrophage-targeting peptide CRVLRSGSC | In vivo (LLC lung cancer in C57BL/6 mice) | Intravenous injection | 198 |
| SIRP-α | Macrophages | RNP | Cationic arginine-coated gold nanoparticles | In vitro (RAW264.7 macrophages) | — | 200 |
| RICTOR | Macrophages | RNP | Liposomes composed of Lipoid S100 and DOTAP | In vivo (4T1 TNBC in BALB/c mice) | Intratumoral injection | 201 |
| YTHDF1 | Dendritic cells | Plasmid | Polymer-LNP (1,2-dioleoyl-3-trimethylammonium-propane and PEG-PLGA) coated with attenuated Salmonella-derived OMVs | In vivo (MC38 colorectal cancer in C57BL/6 mice) | Intravenous injection | 210 |
| PD-1 | T cells | RNP | Liposomes | In vitro (primary T cells from peripheral blood of human) | — | 243 |
3.1. Targeting macrophages
Macrophages exhibit phagocytosis of tumor cells, degradation of dead cells and debris, and modulation of inflammatory processes.185–189 There are two phenotypes of macrophage, where M1 macrophages are usually considered anti-tumoral, pro-inflammatory and immune-activating, whereas M2 phenotype macrophages are related to tumor-associated, anti-inflammatory, and immunosuppressive functions.190–192 Therefore, promotion of the recognition of macrophages of tumor cells and the polarization of macrophage phenotypes from M2 to M1 in tumors can be beneficial for cancer treatment, which can be realized by leveraging genome editing systems. Generally, to enhance the genome editing efficiency in macrophages, non-viral vectors are modified with ligands that can bind to receptors expressed on macrophages, such as macrophage galactose-type lectin and retinoid X receptor beta.193,194Zhao et al.195 developed E. coli protoplast-derived functionalized nanovesicles (NVs) to encapsulate Cas9-sgRNA RNP targeting Pik3cg (also known as PI-3 kinase gamma (PI3Kγ), a critical modulator of macrophage phenotype) (Fig. 7A). The plasmids which encoded sgPik3cg and Cas9 were transformed into E. coli, so the derived NVs exhibited high loading efficiency of the Cas9-sgPik3cg complex. Inactivation of macrophage PI3Kγ led to suppression of the tumor growth by inducing immune activation of the macrophages, and promoted an immunostimulatory transcriptional program that restores CD8+ T cell activation and cytotoxicity.196,197 The NVs were also modified with a pH-responsive PEG-conjugated phospholipid derivative of 1,2-distearoyl-sn-glycero-3-phosphorylethanolamine (DSPE)-hydrazone bond-PEG2000 (DHP) and a TAM-targeted phospholipid derivative of DSPE-galactosamine (DGA) to obtain sgPik3cg-DHP/DGA-NVs, for dual targeting acts towards TAMs. sgPik3cg-DHP/DGA-NV treatment of TAMs in vitro resulted in the significant downregulation of PI3Kγ and macrophage polarization. In the 4T1 breast cancer mouse model, intravenous sgPik3cg-DHP/DGA-NVs treatment exhibited effective macrophage M2-to-M1 polarization and the following TME remodeling from an immunosuppressive “cold” state to an immune active “hot” state.
 | ||
| Fig. 7 Genome editing in macrophages for cancer immunotherapies. (A) Schematic illustration of sgPik3cg-DHP/DGA-NVs for TAM-targeting genome editing for anti-tumor efficacy enhancement.195 (B) The construction of I3E, the mechanism of M1 polarization, and the awakening of the “hot” tumor-immunity.198 | ||
Zhang et al.198 constructed CRVLRSGSC (CRV) peptide-functionalized exosomes to encapsulate the dCas9-KRAB-sgPI3Kγ-encoded plasmid (Fig. 7B). The CRV peptide can selectively home to tumor tissues and target macrophages.194 Additionally, the nuclease-dead mutants of Cas9 (dCas9) fused with a transcription repressor Krüppel-associated box (KRAB) were used for repressing the gene expression with higher efficiency and lower off-target effects.199 The engineered exosomes remodeled the microenvironment with more stimulated M1 macrophages and T cells and decreased percentages of M2 macrophages and MDSCs, leading to the inhibited growth of tumors.
Ray et al.200 established that cationic arginine-coated gold nanoparticles (ArgNPs) can deliver the CRISPR-Cas9 system for knocking out the signal regulatory protein α (SIRP-α) gene in macrophages. The interaction of SIRP-α on macrophages and CD47 on tumor cells sends a “don't eat me” signal, and the elimination of this signal favors the phagocytosis of cancer cells by (−)SIRP-α macrophage. A peptide tag containing glutamic acids (E20-tag) was inserted at the N-terminus of Cas9 protein for efficient cytosolic delivery, and a nuclear localization signal tag was appended at the C terminus to enhance nuclear accumulation. In in vitro experiments, SIRP-α gene knockout exhibited the efficiency of ∼27% in RAW264.7 cells after 48 h incubation, and the SIRP-α knockout RAW264.7 cells revealed a fourfold increase in phagocytosis of cocultured human osteosarcoma cells (U2OS) compared with the un-edited RAW264.7 cells.
Leonard et al.201 used liposomes loaded with CRISPR complex (crRNA and Cas12a) to target rapamycin-insensitive companion of mammalian target of rapamycin (RICTOR), a rapamycin-insensitive companion of mTOR (mammalian target of rapamycin). The deletion of the RICTOR gene in macrophages induces M1 macrophage polarization, because mTORC2 signaling regulates the generation of M2 macrophages.202,203 The Cas12a system is also a member of the CRISPR-associated nucleases that binds and cleaves DNA targets.204,205 CRISPR-RICTOR-liposome successfully knocked down RICTOR expression, leading to M2 macrophage polarization into M1 macrophages in an in vitro 3D TME formed by the co-culture of breast cancer spheres and M2 macrophages.
3.2. Targeting dendritic cells
Dendritic cells are professional antigen-presenting cells that can initiate antigen-specific adaptive immune responses for T cell activation in anti-tumor immunity.206–208 Specifically, dendritic cells recognize the antigens and become maturate, then present the antigens to T cells to trigger the immune system.209 These important steps for anti-tumor immune activation are anticipated to be enhanced through genome editing. Li et al.210 utilized the outer membrane vesicles (OMVs) of bacteria to load CRISPR/Cas9 plasmid (termed BNMs) (Fig. 8). OMVs preserve various bacteria-derived pathogen-associated molecular patterns (PAMPs), which can be processed and presented by dendritic cells.211,212 The CRISPR/Cas9 plasmid was against YTHDF1, whose downregulation in dendritic cells can promote the cross-presentation of tumor antigens and the cross-priming of CD8+ T cells.213 pCas9/gYthdf1 plasmid transfection in vitro in DC2.4 cells resulted in a ∼30% decrease of the Ythdf1 mRNA level. In MC38 tumor-bearing mice, BNMpCas9/gYthdf1 treatment showed 97.72% tumor inhibition compared with the PBS group. In both lymph nodes and spleens after BNMpCas9/gYthdf1 treatment, the numbers of activated CD8+ T cells and memory CD8+ T cells increased sharply, demonstrating the successful activation of CD8+ T cell-mediated antitumor immune cells by Ythdf1 knockout in dendritic cells. | ||
| Fig. 8 Genome editing in dendritic cells for cancer immunotherapies. BNMs specifically target dendritic cells for gene silencing and editing via pathogen recognition.210 (A) In nature, dendritic cells sense and internalize invading pathogens by recognizing various PAMPs, and then digest phagocytosed bacteria into nucleic acids and bacterial antigens to initiate anti-pathogen immunity. (B) The concept of pathogen recognition-driven dendritic cell-specific targeting for functional manipulation. BNMs containing various PAMPs derived from bacteria were prepared for dendritic cell targeting. The various PAMPs of BNMs facilitate the recognition and internalization of BNMs by dendritic cells through the interaction of PAMPs and pattern recognition receptors (PRRs). After endosome escape and dissociation, the siRNA or CRISPR-Cas9 system from BNMs can modulate dendritic cell functions via gene silencing or gene editing. Moreover, the PAMPs of BNMs can stimulate dendritic cell activation via the ligation of PRRs, which is a vital factor for initiating effective antitumor immunity. | ||
3.3. Targeting other innate immune cells
The activated neutrophils provide signals for the activation and maturation of macrophages and dendritic cells.214–216 Thus, neutrophils play a crucial role in regulating immunity during inflammatory conditions.217–220 The CRISPR-Cas9 system has been documented for editing in neutrophils via non-viral vectors to treat diabetes.221 Although currently there is no example of non-viral vector-based genome editing in other innate immune cells by the CRISPR-Cas9 system for cancer immunotherapies, successful delivery of the CRISPR-Cas9 system to these innate immune cells holds promise for cancer immunotherapy in the future. For example, natural killer (NK) cells play an essential role in anti-tumor immunity as the front line in immune surveillance.222,223 However, NK cells are usually dysfunctional in the tumor microenvironment.224 The CRISPR-Cas9 system was applied by electroporation to downregulate the expression of immune checkpoints (e.g., NKG2A) expressed on NK cells for enhanced tumor cell killing efficiency, whereas the employment of non-viral vectors to deliver the CRISPR-Cas system into NK cells for genome editing is still expected.225 In addition, eosinophils display a potent anti-tumorigenic effect.226 Basophils may exhibit anti- or pro-tumorigenic effects, which may be related to the environment and the stage of tumorigenesis.227 MDSCs are immunosuppressive cells, which inhibit the activation of T cells in tumors.228,2294. Genome editing targeting adaptive immune cells
T cells and B cells are the lymphocytes that the adaptive immune system heavily relies on.230 Genome editing in these adaptive immune cells is expected to promote their activation, proliferation, recruitment and infiltration in tumors for activated anti-tumor immunity. In this section, non-viral vector-enabled genome editing in adaptive immune cells (mainly T cells) was introduced (Table 2). Many adaptive immune cells, particularly T cells, are known to be hard to transfect.231 Thus, several strategies were applied to improve the transfection efficiency via non-vectors to T cells, including the application of cell-penetrating peptide to form nanocomplexes, conjugation of T cell targeting ligands, and optimization of the phospholipid and PEG-lipid components in LNPs.232–2344.1. Targeting T cells
CD4+ and CD8+ T cells are essential T lymphocytes in anti-tumor immunity.235,236 CD4+ T cells as the helper T cells can assist the activation of CD8+ T cells, B cells, and other immune cells.237,238 CD8+ T cells as the cytotoxic T cells are responsible for killing tumor cells.239 The activation and infiltration of T cells and their recognition of tumor cells are crucial in T cell-mediated cancer immunotherapy, and these steps can also be the targets for genome editing.240–242 Lu et al.243 loaded RNP in liposomes to knock out the PD-1 gene in T cells. PD-1 knockout T cells showed a significantly higher ability to proliferate, secrete pro-inflammatory cytokine IFN-γ, and kill HepG2 cells than un-edited T cells in vitro. In HepG2 xenografts in mice, edited T cells induced apoptosis of HepG2 cells, leading to significantly suppressed tumor growth and improved mouse survival.4.2. Targeting other adaptive immune cells
B cells include short- or long-lived plasma cells, germinal center cells, and memory cells.244 Although currently a non-viral vector-based CRISPR delivery system has not been applied in genome editing in B cells for cancer immunotherapy, the lipid nanoparticle-mediated delivery of CRISPR pDNA downregulated the number of B cells in vivo for rheumatoid arthritis therapy.245 The successful genome in B cells could also provide possibilities for cancer immunotherapies.5. Conclusions and outlook
Non-viral vectors have emerged as the next-generation delivery system for gene editing systems due to their excellent loading capacity, chemical versatility, and biocompatibility when compared with viral vectors, despite relatively low transfection efficiency in the targeted tissues after administration in vivo. As summarized in this review, utilizing the CRISPR-Cas system to boost cancer immunotherapy has been a promising combination. Non-viral vectors to deliver the CRISPR-Cas system in tumor cells and immune cells have been recently explored, proving the potential in future pre-clinical research. Currently, the immune checkpoints on tumor cells and immune cells (e.g., PD-1 on T cells, PD-L1 on tumor cells) are the common and straightforward targets for non-viral vector-based genome editing for cancer immunotherapy. Besides, some other immune-related genes are also considered as genome editing targets, because their high expression in tumor cells or immune cells can lead to immune cell dysfunction, such as compromised antigen presentation and phagocytosis by immune cells, and macrophage M2-to-M1 polarization disability. Given the fact that these gene expressions in tumors interfere with active anti-tumor immunity, using the CRISPR/Cas system to knock out these genes for immunity retrieval is beneficial.While there is no CRISPR-Cas9 genome editing approved for cancer immunotherapies for clinical applications, it is encouraging that the U.S. Food and Drug Administration (FDA) approved the first CRISPR/Cas9 therapy for treating sickle cell disease in December 2023.246 Clinical trials of the CRISPR-Cas system for cancer therapy have been initiated for targeting leukemia, lymphoma, breast cancer, lung cancer, and other hematologic malignancies and solid tumors, while the employment of non-viral vectors for CRISPR-Cas system delivery for cancer treatments is rare in these clinical trials,42,247,248 which is believed will be seen in the near future.
Despite the great promise, several crucial challenges should be considered and tackled to make non-viral vector-based genome editing more applicable for cancer immunotherapy. First, the safety of the compositions of nanocarriers, including their immunogenicity and biocompatibility, needs to be further ensured. Although lipids, polymers, and inorganic materials were employed to develop the CRISPR-Cas delivery system for successful genome editing and cancer immunotherapy promotion, the FDA-approved materials composition should be taken into more consideration for in vivo genome editing via CRISPR delivery in clinical study. Second, the gene editing efficiency in immune cells could be further improved, which is because targeting immune cells is still challenging, despite the accumulative knowledge about nanoparticles targeting tumor cells. Only a few cases focus on the in vivo targeting of nanocarriers to immune cells rather than in vitro induction. Especially for T cells and B cells, the delivery of CRISPR-Cas systems into these cells is even more difficult, because they are not actively phagocytic cells. Yet, considering the fact that T cells and B cells play important roles in the adaptive anti-immune system and are promising targets for cancer immunotherapy, more effort is expected to validate the targeted delivery of CRISPR based on non-viral carriers to T and B cells. Additionally, non-viral vector-based gene editing in cancer and immune cells can be further combined with other immunotherapeutic strategies (e.g., immune checkpoint blockade and adoptive T cell therapy) or other immune-activating therapies (e.g., radiotherapy, photothermal therapy, photodynamic therapy) for a further enhanced anti-tumor immunotherapeutic efficacy.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the NSERC Discovery Program (RGPIN-2021-02669, to G.C.), NOVA-FRQNT/NSERC Grant (2023-NOVA-314056 & ALLRP 571574-21, to G.C.), and the start-up package of McGill University (to G.C.). T. F. would also like to acknowledge the Rolande and Marcel Gosselin Graduate Studentship, Dr Victor KS Lui Studentship, Charlotte and Leo Karassik Foundation Oncology Ph.D. Fellowship from the Rosalind & Morris Goodman Cancer Institute as well as the BME recruitment award.References
- Z. Liu, M. Shi, Y. Ren, H. Xu, S. Weng, W. Ning, X. Ge, L. Liu, C. Guo, M. Duo, L. Li, J. Li and X. Han, Mol. Cancer, 2023, 22, 35 CrossRef PubMed
.
- S. Tan, D. Li and X. Zhu, Biomed. Pharmacother., 2020, 124, 109821 CrossRef PubMed
.
- S. Farkona, E. P. Diamandis and I. M. Blasutig, BMC Med., 2016, 14, 73 CrossRef PubMed
.
- K. Yi, H. Kong, Y. H. Lao, D. Li, R. L. Mintz, T. Fang, G. Chen, Y. Tao, M. Li and J. Ding, Adv. Mater., 2023, e2300665, DOI:10.1002/adma.202300665
.
- L. Mauge, M. Terme, E. Tartour and D. Helley, Front. Oncol., 2014, 4, 61 Search PubMed
.
- A. Labani-Motlagh, M. Ashja-Mahdavi and A. Loskog, Front. Immunol., 2020, 11, 940 CrossRef CAS PubMed
.
- N. Xie, G. Shen, W. Gao, Z. Huang, C. Huang and L. Fu, Signal Transduction Targeted Ther., 2023, 8, 9 CrossRef CAS PubMed
.
- J. Park, L. Wang and P. C. Ho, Oncogenesis, 2022, 11, 62 CrossRef PubMed
.
- A. Del Prete, V. Salvi, A. Soriani, M. Laffranchi, F. Sozio, D. Bosisio and S. Sozzani, Cell. Mol. Immunol., 2023, 20, 432–447 CrossRef CAS PubMed
.
- S. Y. Wu, T. Fu, Y. Z. Jiang and Z. M. Shao, Mol. Cancer, 2020, 19, 120 CrossRef CAS PubMed
.
- S. Zhao, L. Zhang, S. Xiang, Y. Hu, Z. Wu and J. Shen, Front. Immunol., 2022, 13, 791006 CrossRef CAS PubMed
.
- Y. Liu and G. Zeng, J. Immunother., 2012, 35, 299–308 CrossRef CAS PubMed
.
- D. Li and M. Wu, Signal Transduction Targeted Ther., 2021, 6, 291 CrossRef CAS PubMed
.
- P. Lacy, Front. Immunol., 2015, 6, 190 Search PubMed
.
- G. Arango Duque and A. Descoteaux, Front. Immunol., 2014, 5, 491 Search PubMed
.
- C. N. Baxevanis, Cancers, 2023, 15, 881 CrossRef PubMed
.
- V. Debien, A. De Caluwe, X. Wang, M. Piccart-Gebhart, V. K. Tuohy, E. Romano and L. Buisseret, npj Breast Cancer, 2023, 9, 7 CrossRef PubMed
.
- A. Ribas and J. D. Wolchok, Science, 2018, 359, 1350–1355 CrossRef CAS PubMed
.
- O. Melaiu, V. Lucarini, R. Giovannoni, D. Fruci and F. Gemignani, Semin. Cancer Biol., 2022, 79, 18–43 CrossRef CAS PubMed
.
- E. Baulu, C. Gardet, N. Chuvin and S. Depil, Sci. Adv., 2023, 9, eadf3700 CrossRef CAS PubMed
.
- A. Lahiri, A. Maji, P. D. Potdar, N. Singh, P. Parikh, B. Bisht, A. Mukherjee and M. K. Paul, Mol. Cancer, 2023, 22, 40 CrossRef PubMed
.
- D. M. Pardoll, Nat. Rev. Cancer, 2012, 12, 252–264 CrossRef CAS PubMed
.
- A. Mpakali and E. Stratikos, Cancers, 2021, 13, 134 CrossRef CAS PubMed
.
- Q. Li, Y. Zhou, W. He, X. Ren, M. Zhang, Y. Jiang, Z. Zhou and Y. Luan, J. Controlled Release, 2021, 338, 33–45 CrossRef CAS PubMed
.
- L. Barrueto, F. Caminero, L. Cash, C. Makris, P. Lamichhane and R. R. Deshmukh, Transl. Oncol., 2020, 13, 100738 CrossRef PubMed
.
- J. Pazhani, S. Jayaraman and V. P. Veeraraghavan, Int. J. Surg., 2023, 109, 230–231 CrossRef PubMed
.
- A. Titov, E. Zmievskaya, I. Ganeeva, A. Valiullina, A. Petukhov, A. Rakhmatullina, R. Miftakhova, M. Fainshtein, A. Rizvanov and E. Bulatov, Cancers, 2021, 13, 743 CrossRef CAS PubMed
.
- M. Hiltensperger and A. M. Krackhardt, Front. Immunol., 2023, 14, 1121030 CrossRef CAS PubMed
.
- C. J. Kirk, D. Hartigan-O'Connor and J. J. Mulé, Cancer Res., 2001, 61, 8794–8802 CAS
.
- D. A. Wainwright, A. L. Chang, M. Dey, I. V. Balyasnikova, C. K. Kim, A. Tobias, Y. Cheng, J. W. Kim, J. Qiao, L. Zhang, Y. Han and M. S. Lesniak, Clin. Cancer Res., 2014, 20, 5290–5301 CrossRef CAS PubMed
.
- V. P. Balachandran, G. L. Beatty and S. K. Dougan, Gastroenterology, 2019, 156, 2056–2072 CrossRef CAS PubMed
.
- D. Ciardiello, P. P. Vitiello, C. Cardone, G. Martini, T. Troiani, E. Martinelli and F. Ciardiello, Cancer Treat. Rev., 2019, 76, 22–32 CrossRef CAS PubMed
.
- X. Lu, J. W. Horner, E. Paul, X. Shang, P. Troncoso, P. Deng, S. Jiang, Q. Chang, D. J. Spring, P. Sharma, J. A. Zebala, D. Y. Maeda, Y. A. Wang and R. A. DePinho, Nature, 2017, 543, 728–732 CrossRef CAS PubMed
.
- N. N. Shah and T. J. Fry, Nat. Rev. Clin Oncol., 2019, 16, 372–385 CAS
.
- R. C. Sterner and R. M. Sterner, Blood Cancer J., 2021, 11, 69 CrossRef PubMed
.
- S. Kelderman, T. N. Schumacher and J. B. Haanen, Mol. Oncol., 2014, 8, 1132–1139 CrossRef CAS PubMed
.
- C. M. Jackson, J. Choi and M. Lim, Nat. Immunol., 2019, 20, 1100–1109 CrossRef CAS PubMed
.
- A. Kalbasi and A. Ribas, Nat. Rev. Immunol., 2020, 20, 25–39 CrossRef CAS PubMed
.
- H. Li, Y. Yang, W. Hong, M. Huang, M. Wu and X. Zhao, Signal Transduction Targeted Ther., 2020, 5, 1 CrossRef PubMed
.
- J. C. Carpenter and G. A.-O. Lignani, Neurotherapeutics, 2021, 18, 1515–1523 CrossRef PubMed
.
- K. Liu, J. J. Cui, Y. Zhan, Q. Y. Ouyang, Q. S. Lu, D. H. Yang, X. P. Li and J. Y. Yin, Mol. Cancer, 2022, 21, 98 CrossRef PubMed
.
- Z. Xu, Q. Wang, H. Zhong, Y. Jiang, X. Shi, B. Yuan, N. Yu, S. Zhang, X. Yuan, S. Guo and Y. Yang, Exploration, 2022, 2, 20210081 CrossRef PubMed
.
- Editorial, Nat. Mater., 2021, 20, 1 CrossRef PubMed
.
- H. Zhu, C. Li and C. Gao, Nat. Rev. Mol. Cell Biol., 2020, 21, 661–677 CrossRef CAS PubMed
.
- S. Q. Tsai and J. K. Joung, Nat. Rev. Genet., 2016, 17, 300–312 CrossRef CAS PubMed
.
- T. Fang, X. Cao, M. Ibnat and G. Chen, J. Nanobiotechnol., 2022, 20, 354 CrossRef CAS PubMed
.
- M. Adli, Nat. Commun., 2018, 9, 1911 CrossRef PubMed
.
- T. Li, Y. Yang, H. Qi, W. Cui, L. Zhang, X. Fu, X. He, M. Liu, P. F. Li and T. Yu, Signal Transduction Targeted Ther., 2023, 8, 36 CrossRef PubMed
.
- M. Chavez, X. Chen, P. B. Finn and L. S. Qi, Nat. Rev. Nephrol., 2023, 19, 9–22 CrossRef CAS PubMed
.
- A. Hendel, R. O. Bak, J. T. Clark, A. B. Kennedy, D. E. Ryan, S. Roy, I. Steinfeld, B. D. Lunstad, R. J. Kaiser, A. B. Wilkens, R. Bacchetta, A. Tsalenko, D. Dellinger, L. Bruhn and M. H. Porteus, Nat. Biotechnol., 2015, 33, 985–989 CrossRef CAS PubMed
.
- C. A. Lino, J. C. Harper, J. P. Carney and J. A. Timlin, Drug Delivery, 2018, 25, 1234–1257 CrossRef CAS PubMed
.
- R. M. Gupta and K. Musunuru, J. Clin. Invest., 2014, 124, 4154–4161 CrossRef PubMed
.
- D. J. Segal, B. Dreier, R. R. Beerli and C. F. Barbas, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 2758–2763 CrossRef CAS PubMed
.
- N. E. Sanjana, L. Cong, Y. Zhou, M. M. Cunniff, G. Feng and F. Zhang, Nat. Protoc., 2012, 7, 171–192 CrossRef CAS PubMed
.
- K. Yoshimi and T. Mashimo, J. Hum. Genet., 2018, 63, 115–123 CrossRef CAS PubMed
.
- Z. WareJoncas, J. M. Campbell, G. Martinez-Galvez, W. A. C. Gendron, M. A. Barry, P. C. Harris, C. R. Sussman and S. C. Ekker, Nat. Rev. Nephrol., 2018, 14, 663–677 CrossRef CAS PubMed
.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS PubMed
.
- P. Ma, Q. Wang, X. Luo, L. Mao, Z. Wang, E. Ye, X. J. Loh, Z. Li and Y. L. Wu, Biomater. Sci., 2023, 11, 5078–5094 RSC
.
- A. V. Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, P. J. Chen, C. Wilson, G. A. Newby, A. Raguram and D. R. Liu, Nature, 2019, 576, 149–157 CrossRef CAS PubMed
.
- P. J. Chen and D. R. Liu, Nat. Rev. Genet., 2023, 24, 161–177 CrossRef CAS PubMed
.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Nature, 2017, 551, 464–471 CrossRef CAS PubMed
.
- E. M. Porto, A. C. Komor, I. M. Slaymaker and G. W. Yeo, Nat. Rev. Drug Discovery, 2020, 19, 839–859 CrossRef CAS PubMed
.
- D. Collias, E. Vialetto, J. Yu, K. Co, E. D. H. Almasi, A. S. Ruttiger, T. Achmedov, T. Strowig and C. L. Beisel, Nat. Commun., 2023, 14, 680 CrossRef CAS PubMed
.
- J. K. Lee, E. Jeong, J. Lee, M. Jung, E. Shin, Y. H. Kim, K. Lee, I. Jung, D. Kim, S. Kim and J. S. Kim, Nat. Commun., 2018, 9, 3048 CrossRef PubMed
.
- C. D. Sago, M. P. Lokugamage, K. Paunovska, D. A. Vanover, C. M. Monaco, N. N. Shah, M. Gamboa Castro, S. E. Anderson, T. G. Rudoltz, G. N. Lando, P. Munnilal Tiwari, J. L. Kirschman, N. Willett, Y. C. Jang, P. J. Santangelo, A. V. Bryksin and J. E. Dahlman, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E9944–E9952 CrossRef CAS PubMed
.
- C. Xu, Z. Lu, Y. Luo, Y. Liu, Z. Cao, S. Shen, H. Li, J. Liu, K. Chen, Z. Chen, X. Yang, Z. Gu and J. Wang, Nat. Commun., 2018, 9, 4092 CrossRef PubMed
.
- T. Wei, Q. Cheng, Y. L. Min, E. N. Olson and D. J. Siegwart, Nat. Commun., 2020, 11, 3232 CrossRef CAS PubMed
.
- D. Chaverra-Rodriguez, V. M. Macias, G. L. Hughes, S. Pujhari, Y. Suzuki, D. R. Peterson, D. Kim, S. McKeand and J. L. Rasgon, Nat. Commun., 2018, 9, 3008 CrossRef PubMed
.
- N. Rouatbi, T. McGlynn and K. T. Al-Jamal, Biomater. Sci., 2022, 10, 3410–3432 RSC
.
- Y. Lin, E. Wagner and U. Lachelt, Biomater. Sci., 2022, 10, 1166–1192 RSC
.
- J. O'Keeffe Ahern, I. Lara-Saez, D. Zhou, R. Murillas, J. Bonafont, A. Mencia, M. Garcia, D. Manzanares, J. Lynch, R. Foley, Q. Xu, A. Sigen, F. Larcher and W. Wang, Gene Ther., 2022, 29, 157–170 CrossRef PubMed
.
- D. R. Jacobson and O. A. Saleh, Nucleic Acids Res., 2017, 45, 1596–1605 CAS
.
- S. Zhen and X. Li, Cancer Gene Ther., 2020, 27, 515–527 CrossRef CAS PubMed
.
- C. Zhuo, J. Zhang, J. H. Lee, J. Jiao, D. Cheng, L. Liu, H. W. Kim, Y. Tao and M. Li, Signal Transduction Targeted Ther., 2021, 6, 238 CrossRef CAS PubMed
.
- G. Chen, A. A. Abdeen, Y. Wang, P. K. Shahi, S. Robertson, R. Xie, M. Suzuki, B. R. Pattnaik, K. Saha and S. Gong, Nat. Nanotechnol., 2019, 14, 974–980 CrossRef CAS PubMed
.
- A. P. Chandrasekaran, M. Song, K. S. Kim and S. Ramakrishna, Prog. Mol. Biol. Transl. Sci., 2018, 159, 157–176 CAS
.
- D. Wang, P. W. L. Tai and G. Gao, Nat. Rev. Drug Discovery, 2019, 18, 358–378 CrossRef CAS PubMed
.
- C. Li and R. J. Samulski, Nat. Rev. Genet., 2020, 21, 255–272 CrossRef CAS PubMed
.
- M. Humbel, M. Ramosaj, V. Zimmer, S. Regio, L. Aeby, S. Moser, A. Boizot, M. Sipion, M. Rey and N. Deglon, Gene Ther., 2021, 28, 75–88 CrossRef CAS PubMed
.
- S. E. Ahmadi, M. Soleymani, F. Shahriyary, M. R. Amirzargar, M. Ofoghi, M. D. Fattahi and M. Safa, Cancer Gene Ther., 2023, 1–19, DOI:10.1038/s41417-023-00597-z
.
-
C. E. Nelson, C. L. Duvall, A. Prokop, C. A. Gersbach and J. M. Davidson, in Principles of Tissue Engineering, ed. R. Lanza, R. Langer, J. P. Vacanti and A. Atala, Academic Press, 5th edn, 2020, pp. 519–554. DOI:10.1016/B978-0-12-818422-6.00030-7
.
- F. Uddin, C. M. Rudin and T. Sen, Front. Oncol., 2020, 10, 1387 CrossRef PubMed
.
- M. Ramamoorth and A. Narvekar, J. Clin. Diagn. Res., 2015, 9, GE01-06 Search PubMed
.
- T. Travieso, J. Li, S. Mahesh, J. Mello and M. Blasi, npj Vaccines, 2022, 7, 75 CrossRef CAS PubMed
.
- K. J. Ewer, T. Lambe, C. S. Rollier, A. J. Spencer, A. V. Hill and L. Dorrell, Curr. Opin. Immunol., 2016, 41, 47–54 CrossRef CAS PubMed
.
- C. F. Xu, G. J. Chen, Y. L. Luo, Y. Zhang, G. Zhao, Z. D. Lu, A. Czarna, Z. Gu and J. Wang, Adv. Drug Delivery Rev., 2021, 168, 3–29 CrossRef CAS PubMed
.
- Z. Iqbal, K. Rehman, J. Xia, M. Shabbir, M. Zaman, Y. Liang and L. Duan, Biomater. Sci., 2023, 11, 3762–3783 RSC
.
- Y. Ye, X. Zhang, F. Xie, B. Xu, P. Xie, T. Yang, Q. Shi, C. Y. Zhang, Y. Zhang, J. Chen, X. Jiang and J. Li, Biomater. Sci., 2020, 8, 2966–2976 RSC
.
- Q. Zheng, W. Wang, Y. Zhou, J. Mo, X. Chang, Z. Zha and L. Zha, Biomater. Sci., 2023, 11, 5361–5389 RSC
.
- R. Mout, M. Ray, Y. W. Lee, F. Scaletti and V. M. Rotello, Bioconjugate Chem., 2017, 28, 880–884 CrossRef CAS PubMed
.
- Y. Wang, B. Ma, A. A. Abdeen, G. Chen, R. Xie, K. Saha and S. Gong, ACS Appl. Mater. Interfaces, 2018, 10, 31915–31927 CrossRef CAS PubMed
.
- Z. Glass, Y. Li and Q. Xu, Nat. Biomed. Eng., 2017, 1, 854–855 CrossRef PubMed
.
- H. X. Wang, M. Li, C. M. Lee, S. Chakraborty, H. W. Kim, G. Bao and K. W. Leong, Chem. Rev., 2017, 117, 9874–9906 CrossRef CAS PubMed
.
- T. Wei, Q. Cheng, L. Farbiak, D. G. Anderson, R. Langer and D. J. Siegwart, ACS Nano, 2020, 14, 9243–9262 CrossRef CAS PubMed
.
- G. Chen, B. Ma, Y. Wang and S. Gong, ACS Appl. Mater. Interfaces, 2018, 10, 18515–18523 CrossRef CAS PubMed
.
- J. Eoh and L. Gu, Biomater. Sci., 2019, 7, 1240–1261 RSC
.
- H. Zu and D. Gao, AAPS J., 2021, 23, 78 CrossRef PubMed
.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef CAS PubMed
.
- A. Cifuentes-Rius, A. Desai, D. Yuen, A. P. R. Johnston and N. H. Voelcker, Nat. Nanotechnol., 2021, 16, 37–46 CrossRef CAS PubMed
.
- Z. Zhao, A. Ukidve, J. Kim and S. Mitragotri, Cell, 2020, 181, 151–167 CrossRef CAS PubMed
.
- R. Ephraim, S. Fraser, K. Nurgali and V. Apostolopoulos, Cancers, 2022, 14, 3788 CrossRef PubMed
.
- X. Cai, H. Zhan, Y. Ye, J. Yang, M. Zhang, J. Li and Y. Zhuang, Front. Genet., 2021, 12, 785153 CrossRef CAS PubMed
.
- A. D. Waldman, J. M. Fritz and M. J. Lenardo, Nat. Rev. Immunol., 2020, 20, 651–668 CrossRef CAS PubMed
.
- H. Gonzalez, C. Hagerling and Z. A.-O. Werb.
- P. H. Pandya, M. E. Murray, K. E. Pollok and J. L. Renbarger, J. Immunol. Res., 2016, 2016, 4273943 Search PubMed
.
- G. P. Dunn, C. M. Koebel and R. D. Schreiber, Nat. Rev. Immunol., 2006, 6, 836–848 CrossRef CAS PubMed
.
- C. H. Patel, R. D. Leone, M. R. Horton and J. D. Powell, Nat. Rev. Drug Discovery, 2019, 18, 669–688 CrossRef CAS PubMed
.
- C. N. Baxevanis, S. P. Fortis and S. A. Perez, Semin. Cancer Biol., 2021, 72, 76–89 CrossRef PubMed
.
- Y. Tie, F. Tang, Y. Q. Wei and X. W. Wei, J. Hematol. Oncol., 2022, 15, 61 CrossRef CAS PubMed
.
- J. H. Cha, L. C. Chan, C. W. Li, J. L. Hsu and M. C. Hung, Mol. Cell, 2019, 76, 359–370 CrossRef CAS PubMed
.
- E. I. Buchbinder and A. Desai, Am. J. Clin. Oncol., 2016, 39, 98–106 CrossRef CAS PubMed
.
- T. Powles, J. Walker, J. Andrew Williams and J. Bellmunt, Cancer Treat. Rev., 2020, 82, 101925 CrossRef CAS PubMed
.
- X. Wang, F. Teng, L. Kong and J. Yu, OncoTargets Ther., 2016, 9, 5023–5039 CrossRef CAS PubMed
.
- C. J. Li, L. T. Lin, M. F. Hou and P. Y. Chu, Oncol. Rep., 2021, 45, 5–12 CrossRef PubMed
.
- J. Yang, M. Dong, Y. Shui, Y. Zhang, Z. Zhang, Y. Mi, X. Zuo, L. Jiang, K. Liu, Z. Liu, X. Gu and Y. Shi, Cancer Cell Int., 2020, 20, 96 CrossRef CAS PubMed
.
- N. Yaghoubi, A. Soltani, K. Ghazvini, S. M. Hassanian and S. I. Hashemy, Biomed. Pharmacother., 2019, 110, 312–318 CrossRef CAS PubMed
.
- W. J. Cheng, L. C. Chen, H. O. Ho, H. L. Lin and M. T. Sheu, Int. J. Nanomed., 2018, 13, 7079–7094 CrossRef CAS PubMed
.
- A. H. Sharpe and K. E. Pauken, Nat. Rev. Immunol., 2018, 18, 153–167 CrossRef CAS PubMed
.
- F. K. Dermani, P. Samadi, G. Rahmani, A. K. Kohlan and R. Najafi, J. Cell Physiol., 2019, 234, 1313–1325 CrossRef CAS PubMed
.
- C. Sun, R. Mezzadra and T. N. Schumacher, Immunity, 2018, 48, 434–452 CrossRef CAS PubMed
.
- X. He and C. Xu, Cell Res., 2020, 30, 660–669 CrossRef PubMed
.
- H. Deng, S. Tan, X. Gao, C. Zou, C. Xu, K. Tu, Q. Song, F. Fan, W. Huang and Z. Zhang, Acta Pharm. Sin. B, 2020, 10, 358–373 CrossRef CAS PubMed
.
- Y. Lu, F. Wu, Y. Xu, C. He, S. Luo and X. Sun, J. Controlled Release, 2023, 354, 57–68 CrossRef CAS PubMed
.
- Q. Liu, C. Wang, Y. Zheng, Y. Zhao, Y. Wang, J. Hao, X. Zhao, K. Yi, L. Shi, C. Kang and Y. Liu, Biomaterials, 2020, 258, 120275 CrossRef CAS PubMed
.
- Y. Xing, J. Yang, Y. Wang, C. Wang, Z. Pan, F. L. Liu, Y. Liu and Q. Liu, ACS Nano, 2023, 17, 5713–5726 CrossRef CAS PubMed
.
- S. Deshayes, H. Cabral, T. Ishii, Y. Miura, S. Kobayashi, T. Yamashita, A. Matsumoto, Y. Miyahara, N. Nishiyama and K. Kataoka, J. Am. Chem. Soc., 2013, 135, 15501–15507 CrossRef CAS PubMed
.
- J. M. M. Levy, C. G. Towers and A. Thorburn, Nat. Rev. Cancer, 2017, 17, 528–542 CrossRef CAS PubMed
.
- J. Zhou, G. Wang, Y. Chen, H. Wang, Y. Hua and Z. Cai, J. Cell. Mol. Med., 2019, 23, 4854–4865 CrossRef PubMed
.
- J. Liu, G. Li, H. Guo, C. Ni, Y. Gao, X. Cao, J. Xia, X. Shi and R. Guo, ACS Appl. Mater. Interfaces, 2023, 15, 12809–12821 CrossRef CAS PubMed
.
- Y. Lin, U. Wilk, J. Pohmerer, E. Horterer, M. Hohn, X. Luo, H. Mai, E. Wagner and U. Lachelt, Small, 2023, 19, e2205318 CrossRef PubMed
.
- P. M. Klein, S. Kern, D. J. Lee, J. Schmaus, M. Hohn, J. Gorges, U. Kazmaier and E. Wagner, Biomaterials, 2018, 178, 630–642 CrossRef CAS PubMed
.
- E. Y. Chiang and I. Mellman, J. Immunother. Cancer, 2022, 10, e004711 CrossRef PubMed
.
- H. Stamm, F. Klingler, E. M. Grossjohann, J. Muschhammer, E. Vettorazzi, M. Heuser, U. Mock, F. Thol, G. Vohwinkel, E. Latuske, C. Bokemeyer, R. Kischel, C. Dos Santos, S. Stienen, M. Friedrich, M. Lutteropp, D. Nagorsen, J. Wellbrock and W. Fiedler, Oncogene, 2018, 37, 5269–5280 CrossRef CAS PubMed
.
- C. Yue, S. Gao, S. Li, Z. Xing, H. Qian, Y. Hu, W. Wang and C. Hua, Front. Immunol., 2022, 13, 911919 CrossRef CAS PubMed
.
- J. Yeo, M. Ko, D. H. Lee, Y. Park and H. S. Jin, Pharmaceuticals, 2021, 14, 200 CrossRef CAS PubMed
.
- H. Stamm, L. Oliveira-Ferrer, E. M. Grossjohann, J. Muschhammer, V. Thaden, F. Brauneck, R. Kischel, V. Muller, C. Bokemeyer, W. Fiedler and J. Wellbrock, OncoImmunology, 2019, 8, e1674605 CrossRef PubMed
.
- Z. Zhang, Q. Wang, Q. Liu, Y. Zheng, C. Zheng, K. Yi, Y. Zhao, Y. Gu, Y. Wang, C. Wang, X. Zhao, L. Shi, C. Kang and Y. Liu, Adv. Mater., 2019, 31, e1905751 CrossRef PubMed
.
- J. Tao, D. E. Bauer and R. Chiarle, Nat. Commun., 2023, 14, 212 CrossRef CAS PubMed
.
- F. Bigini, S. H. Lee, Y. J. Sun, Y. Sun and V. B. Mahajan, Vision Res., 2023, 213, 108317 CrossRef PubMed
.
- M. Damaghi, N. K. Tafreshi, M. C. Lloyd, R. Sprung, V. Estrella, J. W. Wojtkowiak, D. L. Morse, J. M. Koomen, M. M. Bui, R. A. Gatenby and R. J. Gillies, Nat. Commun., 2015, 6, 8752 CrossRef CAS PubMed
.
- B. Perillo, M. Di Donato, A. Pezone, E. Di Zazzo, P. Giovannelli, G. Galasso, G. Castoria and A. Migliaccio, Exp. Mol. Med., 2020, 52, 192–203 CrossRef CAS PubMed
.
- D. Zhang, G. Wang, X. Yu, T. Wei, L. Farbiak, L. T. Johnson, A. M. Taylor, J. Xu, Y. Hong, H. Zhu and D. J. Siegwart, Nat. Nanotechnol., 2022, 17, 777–787 CrossRef CAS PubMed
.
- M. S. Bauer, F. Baumann, C. Daday, P. Redondo, E. Durner, M. A. Jobst, L. F. Milles, D. Mercadante, D. A. Pippig, H. E. Gaub, F. Grater and D. Lietha, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 6766–6774 CrossRef CAS PubMed
.
- H. Jiang, S. Hegde, B. L. Knolhoff, Y. Zhu, J. M. Herndon, M. A. Meyer, T. M. Nywening, W. G. Hawkins, I. M. Shapiro, D. T. Weaver, J. A. Pachter, A. Wang-Gillam and D. G. DeNardo, Nat. Med., 2016, 22, 851–860 CrossRef CAS PubMed
.
- A. Serrels, T. Lund, B. Serrels, A. Byron, R. C. McPherson, A. von Kriegsheim, L. Gomez-Cuadrado, M. Canel, M. Muir, J. E. Ring, E. Maniati, A. H. Sims, J. A. Pachter, V. G. Brunton, N. Gilbert, S. M. Anderton, R. J. Nibbs and M. C. Frame, Cell, 2015, 163, 160–173 CrossRef CAS PubMed
.
- M. P. Chao, C. H. Takimoto, D. D. Feng, K. McKenna, P. Gip, J. Liu, J. P. Volkmer, I. L. Weissman and R. Majeti, Front. Oncol., 2019, 9, 1380 CrossRef PubMed
.
- R. H. Vonderheide, Nat. Med., 2015, 21, 1122–1123 CrossRef CAS PubMed
.
- Y. Liu, Y. Wang, Y. Yang, L. Weng, Q. Wu, J. Zhang, P. Zhao, L. Fang, Y. Shi and P. Wang, Signal Transduction Targeted Ther., 2023, 8, 104 CrossRef CAS PubMed
.
- Z. Bian, L. Shi, K. Kidder, K. Zen, C. Garnett-Benson and Y. Liu, Nat. Commun., 2021, 12, 3229 CrossRef CAS PubMed
.
- M. Zhang, G. Hutter, S. A. Kahn, T. D. Azad, S. Gholamin, C. Y. Xu, J. Liu, A. S. Achrol, C. Richard, P. Sommerkamp, M. K. Schoen, M. N. McCracken, R. Majeti, I. Weissman, S. S. Mitra and S. H. Cheshier, PLoS One, 2016, 11, e0153550 CrossRef PubMed
.
- M. A. Morrissey, N. Kern and R. D. Vale, Immunity, 2020, 53, 290–302 CrossRef CAS PubMed
.
- Y. Kojima, J. P. Volkmer, K. McKenna, M. Civelek, A. J. Lusis, C. L. Miller, D. Direnzo, V. Nanda, J. Ye, A. J. Connolly, E. E. Schadt, T. Quertermous, P. Betancur, L. Maegdefessel, L. P. Matic, U. Hedin, I. L. Weissman and N. J. Leeper, Nature, 2016, 536, 86–90 CrossRef CAS PubMed
.
- M. Feng, W. Jiang, B. Y. S. Kim, C. C. Zhang, Y. X. Fu and I. L. Weissman, Nat. Rev. Cancer, 2019, 19, 568–586 CrossRef CAS PubMed
.
- Q. Huang, M. Su, L. Zhao, Z. Zhang, Y. Zhang, X. Yang and J. Wang, Nano Today, 2023, 50, 101857 CrossRef CAS
.
- M. Lin, Z. Yang, Y. Yang, Y. Peng, J. Li, Y. Du, Q. Sun, D. Gao, Q. Yuan, Y. Zhou, X. Chen and X. Qi, Nano Today, 2022, 42, 101359 CrossRef CAS
.
- F. Ren, R. Tang, X. Zhang, W. M. Madushi, D. Luo, Y. Dang, Z. Li, K. Wei and G. Chen, PLoS One, 2015, 10, e0135544 CrossRef PubMed
.
- K. Mikecz, F. R. Brennan, J. H. Kim and T. T. Glant, Nat. Med., 1995, 1, 558–563 CrossRef CAS PubMed
.
- P. Kesharwani, R. Chadar, A. Sheikh, W. Y. Rizg and A. Y. Safhi, Front. Pharmacol., 2021, 12, 800481 CrossRef CAS PubMed
.
- Y. Xing, J. Yang, C. Wang, Z. Kang, Z. Pan, J. Tang, F. Li, X. Wang, X.-m. Meng, Z. Cheng, Y. Liu and Q. Liu, Chem. Eng. J., 2023, 474, 145796 CrossRef CAS
.
- J. Wei, L. Long, W. Zheng, Y. Dhungana, S. A. Lim, C. Guy, Y. Wang, Y. D. Wang, C. Qian, B. Xu, A. Kc, J. Saravia, H. Huang, J. Yu, J. G. Doench, T. L. Geiger and H. Chi, Nature, 2019, 576, 471–476 CrossRef CAS PubMed
.
- T. Xia, X. M. Yi, X. Wu, J. Shang and H. B. Shu, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 20063–20069 CrossRef CAS PubMed
.
- P. F. Wang, H. Q. Cai, C. B. Zhang, Y. M. Li, X. Liu, J. H. Wan, T. Jiang, S. W. Li and C. X. Yan, J. Neuroinflammation, 2018, 15, 145 CrossRef PubMed
.
- Z. Huang, M. Liu, D. Li, Y. Tan, R. Zhang, Z. Xia, P. Wang, B. Jiao, P. Liu and R. Ren, J. Biol. Chem., 2020, 295, 18343–18354 CrossRef CAS PubMed
.
- R. T. Manguso, H. W. Pope, M. D. Zimmer, F. D. Brown, K. B. Yates, B. C. Miller, N. B. Collins, K. Bi, M. W. LaFleur, V. R. Juneja, S. A. Weiss, J. Lo, D. E. Fisher, D. Miao, E. Van Allen, D. E. Root, A. H. Sharpe, J. G. Doench and W. N. Haining, Nature, 2017, 547, 413–418 CrossRef CAS PubMed
.
- M. W. LaFleur, T. H. Nguyen, M. A. Coxe, B. C. Miller, K. B. Yates, J. E. Gillis, D. R. Sen, E. F. Gaudiano, R. Al Abosy, G. J. Freeman, W. N. Haining and A. H. Sharpe, Nat. Immunol., 2019, 20, 1335–1347 CrossRef CAS PubMed
.
- M. W. LaFleur, T. H. Nguyen, M. A. Coxe, K. B. Yates, J. D. Trombley, S. A. Weiss, F. D. Brown, J. E. Gillis, D. J. Coxe, J. G. Doench, W. N. Haining and A. H. Sharpe, Nat. Commun., 2019, 10, 1668 CrossRef PubMed
.
- F. Wiede, K. H. Lu, X. Du, S. Liang, K. Hochheiser, G. T. Dodd, P. K. Goh, C. Kearney, D. Meyran, P. A. Beavis, M. A. Henderson, S. L. Park, J. Waithman, S. Zhang, Z. Y. Zhang, J. Oliaro, T. Gebhardt, P. K. Darcy and T. Tiganis, EMBO J., 2020, 39, e103637 CrossRef CAS PubMed
.
- C. Yang, Y. Fu, C. Huang, D. Hu, K. Zhou, Y. Hao, B. Chu, Y. Yang and Z. Qian, Biomaterials, 2020, 255, 120194 CrossRef CAS PubMed
.
- J. Yang, K. Yang, S. Du, W. Luo, C. Wang, H. Liu, K. Liu, Z. Zhang, Y. Gao, X. Han and Y. Song, Angew. Chem., Int. Ed., 2023, 62, e202306863 CrossRef CAS PubMed
.
- D. G. Roy, J. Chen, V. Mamane, E. H. Ma, B. M. Muhire, R. D. Sheldon, T. Shorstova, R. Koning, R. M. Johnson, E. Esaulova, K. S. Williams, S. Hayes, M. Steadman, B. Samborska, A. Swain, A. Daigneault, V. Chubukov, T. P. Roddy, W. Foulkes, J. A. Pospisilik, M. C. Bourgeois-Daigneault, M. N. Artyomov, M. Witcher, C. M. Krawczyk, C. Larochelle and R. G. Jones, Cell Metab., 2020, 31, 250–266 CrossRef CAS PubMed
.
- P. M. Gubser and A. Kallies, Immunol. Cell Biol., 2020, 98, 623–625 CrossRef CAS PubMed
.
- Y. Huang, G. Qin, T. Cui, C. Zhao, J. Ren and X. Qu, Nat. Commun., 2023, 14, 4647 CrossRef CAS PubMed
.
- A. Rosati, A. Basile, R. D'Auria, M. d'Avenia, M. De Marco, A. Falco, M. Festa, L. Guerriero, V. Iorio, R. Parente, M. Pascale, L. Marzullo, R. Franco, C. Arra, A. Barbieri, D. Rea, G. Menichini, M. Hahne, M. Bijlsma, D. Barcaroli, G. Sala, F. F. di Mola, P. di Sebastiano, J. Todoric, L. Antonucci, V. Corvest, A. Jawhari, M. A. Firpo, D. A. Tuveson, M. Capunzo, M. Karin, V. De Laurenzi and M. C. Turco, Nat. Commun., 2015, 6, 8695 CrossRef CAS PubMed
.
- L. Marzullo, M. C. Turco and M. De Marco, Biochim Biophys Acta Gen Subj, 2020, 1864, 129628 CrossRef CAS PubMed
.
- M. F. Romano, M. Festa, G. Pagliuca, R. Lerose, R. Bisogni, F. Chiurazzi, G. Storti, S. Volpe, S. Venuta, M. C. Turco and A. Leone, Cell Death Differ., 2003, 10, 383–385 CrossRef CAS PubMed
.
- T. A. Colvin, V. L. Gabai, J. Gong, S. K. Calderwood, H. Li, S. Gummuluru, O. N. Matchuk, S. G. Smirnova, N. V. Orlova, I. A. Zamulaeva, M. Garcia-Marcos, X. Li, Z. T. Young, J. N. Rauch, J. E. Gestwicki, S. Takayama and M. Y. Sherman, Cancer Res., 2014, 74, 4731–4740 CrossRef CAS PubMed
.
- A. Rosati, V. Graziano, V. De Laurenzi, M. Pascale and M. C. Turco, Cell Death Dis., 2011, 2, e141 CrossRef CAS PubMed
.
- B. K. Wang, X. F. Yu, J. H. Wang, Z. B. Li, P. H. Li, H. Wang, L. Song, P. K. Chu and C. Li, Biomaterials, 2016, 78, 27–39 CrossRef CAS PubMed
.
- J. Joung, P. C. Kirchgatterer, A. Singh, J. H. Cho, S. P. Nety, R. C. Larson, R. K. Macrae, R. Deasy, Y. Y. Tseng, M. V. Maus and F. Zhang, Nat. Commun., 2022, 13, 1606 CrossRef CAS PubMed
.
- L. Avinery, V. Gahramanov, A. Hesin and M. Y. Sherman, Cancers, 2022, 14, 4168 CrossRef CAS PubMed
.
- M. De Marco, A. Falco, R. Iaccarino, A. Raffone, A. Mollo, M. Guida, A. Rosati, M. Chetta, G. Genovese, F. De Caro, M. Capunzo, M. C. Turco, V. N. Uversky and L. Marzullo, Br. J. Cancer, 2021, 125, 789–797 CrossRef CAS PubMed
.
- M. De Marco, M. C. Turco and L. Marzullo, Trends Cancer, 2020, 6, 985–988 CrossRef CAS PubMed
.
- X. Chen, S. Wang, Y. Chen, H. Xin, S. Zhang, D. Wu, Y. Xue, M. Zha, H. Li, K. Li, Z. Gu, W. Wei and Y. Ping, Nat. Nanotechnol., 2023, 18, 933–944 CrossRef CAS PubMed
.
- E. B. Okeke and J. E. Uzonna, Front. Immunol., 2019, 10, 680 CrossRef CAS PubMed
.
-
C. J. Burrell, C. R. Howard and F. A. Murphy, in Fenner and White's Medical Virology, 2017, pp. 57–64. DOI:10.1016/b978-0-12-375156-0.00005-9
.
- A. Hochreiter-Hufford and K. S. Ravichandran, Cold Spring Harbor Perspect. Biol., 2013, 5, a008748 CrossRef PubMed
.
- P. Mehrotra and K. S. Ravichandran, Nat. Rev. Drug Discovery, 2022, 21, 601–620 CrossRef CAS PubMed
.
- S. Arandjelovic and K. S. Ravichandran, Nat. Immunol., 2015, 16, 907–917 CrossRef CAS PubMed
.
- C. Varol, A. Mildner and S. Jung, Annu. Rev. Immunol., 2015, 33, 643–675 CrossRef CAS PubMed
.
- S. Kadomoto, K. Izumi and A. Mizokami, Int. J. Mol. Sci., 2022, 23, 144 CrossRef CAS PubMed
.
- J. Liu, X. Geng, J. Hou and G. Wu, Cancer Cell Int., 2021, 21, 389 CrossRef CAS PubMed
.
- Z. Duan and Y. Luo, Signal Transduction Targeted Ther., 2021, 6, 127 CrossRef CAS PubMed
.
- A. Gabba, A. Bogucka, J. G. Luz, A. Diniz, H. Coelho, F. Corzana, F. J. Canada, F. Marcelo, P. V. Murphy and G. Birrane, Biochemistry, 2021, 60, 1327–1336 CrossRef CAS PubMed
.
- T. Tang, Y. Wei, J. Kang, Z. G. She, D. Kim, M. J. Sailor, E. Ruoslahti and H. B. Pang, J. Controlled Release, 2019, 301, 42–53 CrossRef CAS PubMed
.
- M. Zhao, X. Cheng, P. Shao, Y. Dong, Y. Wu, L. Xiao, Z. Cui, X. Sun, C. Gao, J. Chen, Z. Huang and J. Zhang, Nat. Commun., 2024, 15, 950 CrossRef CAS PubMed
.
- M. M. Kaneda, K. S. Messer, N. Ralainirina, H. Li, C. J. Leem, S. Gorjestani, G. Woo, A. V. Nguyen, C. C. Figueiredo, P. Foubert, M. C. Schmid, M. Pink, D. G. Winkler, M. Rausch, V. J. Palombella, J. Kutok, K. McGovern, K. A. Frazer, X. Wu, M. Karin, R. Sasik, E. E. Cohen and J. A. Varner, Nature, 2016, 539, 437–442 CrossRef CAS PubMed
.
- X. Xiang, J. Wang, D. Lu and X. Xu, Signal Transduction Targeted Ther., 2021, 6, 75 CrossRef PubMed
.
- L. Zhang, Y. Lin, S. Li, X. Guan and X. Jiang, Angew. Chem., Int. Ed., 2023, 62, e202217089 CrossRef CAS PubMed
.
- L. S. Qi, M. H. Larson, L. A. Gilbert, J. A. Doudna, J. S. Weissman, A. P. Arkin and W. A. Lim, Cell, 2013, 152, 1173–1183 CrossRef CAS PubMed
.
- M. Ray, Y. W. Lee, J. Hardie, R. Mout, G. Yesilbag Tonga, M. E. Farkas and V. M. Rotello, Bioconjugate Chem., 2018, 29, 445–450 CrossRef CAS PubMed
.
- F. Leonard, L. T. Curtis, A. R. Hamed, C. Zhang, E. Chau, D. Sieving, B. Godin and H. B. Frieboes, Cancer Immunol. Immunother., 2020, 69, 731–744 CrossRef CAS PubMed
.
- R. W. Hallowell, S. L. Collins, J. M. Craig, Y. Zhang, M. Oh, P. B. Illei, Y. Chan-Li, C. L. Vigeland, W. Mitzner, A. L. Scott, J. D. Powell and M. R. Horton, Nat. Commun., 2017, 8, 14208 CrossRef CAS PubMed
.
- W. T. Festuccia, P. Pouliot, I. Bakan, D. M. Sabatini and M. Laplante, PLoS One, 2014, 9, e95432 CrossRef PubMed
.
- D. C. Swarts and M. Jinek, Wiley Interdiscip. Rev.: RNA, 2018, 9, e1481 CrossRef PubMed
.
- A. R. Krysler, C. R. Cromwell, T. Tu, J. Jovel and B. P. Hubbard, Nat. Commun., 2022, 13, 1617 CrossRef CAS PubMed
.
- F. Alfei, P. C. Ho and W. L. Lo, Oncogene, 2021, 40, 5253–5261 CrossRef CAS PubMed
.
- R. M. Steinman, Annu. Rev. Immunol., 1991, 9, 271–296 CrossRef CAS PubMed
.
- C. Fu and A. Jiang, Front. Immunol., 2018, 9, 3059 CrossRef CAS PubMed
.
- Y. Wang, Y. Xiang, V. W. Xin, X. W. Wang, X. C. Peng, X. Q. Liu, D. Wang, N. Li, J. T. Cheng, Y. N. Lyv, S. Z. Cui, Z. Ma, Q. Zhang and H. W. Xin, J. Hematol. Oncol., 2020, 13, 107 CrossRef CAS PubMed
.
- M. Li, H. Zhou, N. Wu, W. Deng, W. Dong, X. Sun, H. Liu, Z. Tian and Y. Wang, Nano Lett., 2023, 23, 2733–2742 CrossRef CAS PubMed
.
- M. G. Sartorio, E. J. Pardue, M. F. Feldman and M. F. Haurat, Annu. Rev. Microbiol., 2021, 75, 609–630 CrossRef CAS PubMed
.
- K. Cheng, R. Zhao, Y. Li, Y. Qi, Y. Wang, Y. Zhang, H. Qin, Y. Qin, L. Chen, C. Li, J. Liang, Y. Li, J. Xu, X. Han, G. J. Anderson, J. Shi, L. Ren, X. Zhao and G. Nie, Nat. Commun., 2021, 12, 2041 CrossRef CAS PubMed
.
- D. Han, J. Liu, C. Chen, L. Dong, Y. Liu, R. Chang, X. Huang, Y. Liu, J. Wang, U. Dougherty, M. B. Bissonnette, B. Shen, R. R. Weichselbaum, M. M. Xu and C. He, Nature, 2019, 566, 270–274 CrossRef CAS PubMed
.
- I. Puga, M. Cols and A. Cerutti, Inmunologia, 2013, 32, 25–34 CrossRef
.
- J. A. Marwick, R. Mills, O. Kay, K. Michail, J. Stephen, A. G. Rossi, I. Dransfield and N. Hirani, Cell Death Dis., 2018, 9, 665 CrossRef PubMed
.
- M. Yi, T. Li, M. Niu, Q. Mei, B. Zhao, Q. Chu, Z. Dai and K. Wu, Mol. Cancer, 2023, 22, 187 CrossRef CAS PubMed
.
- E. Mortaz, S. D. Alipoor, I. M. Adcock, S. Mumby and L. Koenderman, Front. Immunol., 2018, 9, 2171 CrossRef PubMed
.
- C. Rosales, Front. Physiol., 2018, 9, 113 CrossRef PubMed
.
- V. Kumar and A. Sharma, Int. Immunopharmacol., 2010, 10, 1325–1334 CrossRef CAS PubMed
.
- A. Herrero-Cervera, O. Soehnlein and E. Kenne, Cell. Mol. Immunol., 2022, 19, 177–191 CrossRef CAS PubMed
.
- Y. Liu, Z. T. Cao, C. F. Xu, Z. D. Lu, Y. L. Luo and J. Wang, Biomaterials, 2018, 172, 92–104 CrossRef CAS PubMed
.
- T. J. Laskowski, A. Biederstadt and K. Rezvani, Nat. Rev. Cancer, 2022, 22, 557–575 CrossRef CAS PubMed
.
- E. Vivier, E. Tomasello, M. Baratin, T. Walzer and S. Ugolini, Nat. Immunol., 2008, 9, 503–510 CrossRef CAS PubMed
.
- J. Bi and Z. Tian, Front. Immunol., 2017, 8, 760 CrossRef PubMed
.
- T. Bexte, J. Alzubi, L. M. Reindl, P. Wendel, R. Schubert, E. Salzmann-Manrique, I. von Metzler, T. Cathomen and E. Ullrich, OncoImmunology, 2022, 11, 2081415 CrossRef PubMed
.
- S. Ghaffari and N. Rezaei, J. Transl. Med., 2023, 21, 551 CrossRef CAS PubMed
.
-
G. Marone, A. R. Gambardella, F. Mattei, J. Mancini, G. Schiavoni and G. Varricchi, in Tumor Microenvironment: Hematopoietic Cells – Part A, ed. A. Birbrair, Springer International Publishing, Cham, 2020, pp. 21–34. DOI:10.1007/978-3-030-35723-8_2
.
- M. K. Srivastava, P. Sinha, V. K. Clements, P. Rodriguez and S. Ostrand-Rosenberg, Cancer Res., 2010, 70, 68–77 CrossRef CAS PubMed
.
- M. Rowlands, F. Segal and D. Hartl, Front. Immunol., 2021, 12, 697405 CrossRef CAS PubMed
.
- J. S. Marshall, R. Warrington, W. Watson and H. L. Kim, Allergy, Asthma, Clin. Immunol., 2018, 14, 49 CrossRef PubMed
.
- I. Rahimmanesh, M. Totonchi and H. Khanahmad, Res. Pharm. Sci., 2020, 15, 437–446 CrossRef PubMed
.
- D. V. Foss, J. J. Muldoon, D. N. Nguyen, D. Carr, S. U. Sahu, J. M. Hunsinger, S. K. Wyman, N. Krishnappa, R. Mendonsa, E. V. Schanzer, B. R. Shy, V. S. Vykunta, V. Allain, Z. Li, A. Marson, J. Eyquem and R. C. Wilson, Nat. Biomed. Eng., 2023, 7, 647–660 CrossRef CAS PubMed
.
- A. Kheirolomoom, A. J. Kare, E. S. Ingham, R. Paulmurugan, E. R. Robinson, M. Baikoghli, M. Inayathullah, J. W. Seo, J. Wang, B. Z. Fite, B. Wu, S. K. Tumbale, M. N. Raie, R. H. Cheng, L. Nichols, A. D. Borowsky and K. W. Ferrara, Biomaterials, 2022, 281, 121339 CrossRef CAS PubMed
.
- H. Tanaka, R. Miyama, Y. Sakurai, S. Tamagawa, Y. Nakai, K. Tange, H. Yoshioka and H. Akita, Pharmaceutics, 2021, 13, 2097 CrossRef CAS PubMed
.
- V. Appay, R. A. van Lier, F. Sallusto and M. Roederer, Cytometry, Part A, 2008, 73, 975–983 CrossRef PubMed
.
- D. N. Nguyen, T. L. Roth, P. J. Li, P. A. Chen, R. Apathy, M. R. Mamedov, L. T. Vo, V. R. Tobin, D. Goodman, E. Shifrut, J. A. Bluestone, J. M. Puck, F. C. Szoka and A. Marson, Nat. Biotechnol., 2020, 38, 44–49 CrossRef CAS PubMed
.
- S. L. Swain, K. K. McKinstry and T. M. Strutt, Nat. Rev. Immunol., 2012, 12, 136–148 CrossRef CAS PubMed
.
- P. Castiglioni, M. Gerloni, X. Cortez-Gonzalez and M. Zanetti, Eur. J. Immunol., 2005, 35, 1360–1370 CrossRef CAS PubMed
.
- A. Durgeau, Y. Virk, S. Corgnac and F. Mami-Chouaib, Front. Immunol., 2018, 9, 14 CrossRef PubMed
.
- M. Wang, H. Y. Yin, B. Fau-Wang, R.-F. Wang Hy Fau-Wang and R. F. Wang, Front. Immunol., 2018, 9, 14 CrossRef PubMed
.
- B. Weigelin and P. Friedl, Trends Cancer, 2022, 8, 980–987 CrossRef CAS PubMed
.
- M. J. W. Sim and P. D. Sun, Front. Immunol., 2022, 13, 833017 CrossRef CAS PubMed
.
- S. Lu, N. Yang, J. He, W. Gong, Z. Lai, L. Xie, L. Tao, C. Xu, H. Wang, G. Zhang, H. Cao, C. Zhou, L. Zhong and Y. Zhao, J. Biomed. Nanotechnol., 2019, 15, 593–601 CrossRef CAS PubMed
.
- J. G. Cyster and C. D. C. Allen, Cell, 2019, 177, 524–540 CrossRef CAS PubMed
.
- M. Li, Y.-N. Fan, Z.-Y. Chen, Y.-L. Luo, Y.-C. Wang, Z.-X. Lian, C.-F. Xu and J. Wang, Nano Res., 2018, 11, 6270–6282 CrossRef CAS
.
- M. Senior, Nat. Biotechnol., 2024, 42, 355–361 CrossRef CAS PubMed
.
- H. Kong, E. Ju, K. Yi, W. Xu, Y. H. Lao, D. Cheng, Q. Zhang, Y. Tao, M. Li and J. Ding, Adv. Sci., 2021, 8, e2102051 CrossRef PubMed
.
- S. Zhang, Y. Wang, D. Mao, Y. Wang, H. Zhang, Y. Pan, Y. Wang, S. Teng and P. Huang, Front. Med., 2023, 10, 1292452 CrossRef PubMed
.
| This journal is © The Royal Society of Chemistry 2024 |

