Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Two-dimensional molybdenum disulfide nanosheets evoke nitric oxide-dependent antibacterial effects†
Guotao
Peng
 ab,
Viviana
González
ab,
Viviana
González
 c,
Ester
Vázquez
c,
Ester
Vázquez
 cd,
Jon O.
Lundberg
e and
Bengt
Fadeel
cd,
Jon O.
Lundberg
e and
Bengt
Fadeel
 *a
*a
aInstitute of Environmental Medicine, Karolinska Institutet, Stockholm, Sweden. E-mail: bengt.fadeel@ki.se
bCollege of Environmental Science and Engineering, Tongji University, Shanghai, China
cInstituto Regional de Investigación Científica Aplicada, Universidad de Castilla-La Mancha, Ciudad Real, Spain
dFacultad de Ciencias y Teconologías Químicas, Universidad de Castilla-La Mancha, Ciudad Real, Spain
eDepartment of Physiology and Pharmacology, Karolinska Institutet, Stockholm, Sweden
First published on 10th October 2023
Abstract
Nanomaterials are currently being explored as novel antimicrobial agents. In this study, we first investigated the ability of two-dimensional (2D) molybdenum disulfide (MoS2) nanosheets to trigger neutrophil extracellular traps (NETs) using neutrophil-differentiated HL-60 cells as well as primary human peripheral blood neutrophils. We then addressed whether the MoS2 nanosheets themselves function as antibacterial agents. We found that MoS2 and Na2MoO4 both triggered NETs, as evidenced by the quantification of neutrophil elastase (NE) activity and immunofluorescence staining of extracellular NE, as well as scanning electron microscopy. The release of NETs was found to be nitric oxide (NO)-dependent. We also found that the MoS2 nanosheets but not the soluble salt prompted acellular NO production in the presence of NaNO2. The acellular generation of NO, suggestive of nanozyme properties of the MoS2 nanosheets, was demonstrated by electron paramagnetic resonance analysis. Electrochemical analysis using cyclic voltammetry confirmed the redox transition of the MoS2 nanosheets. Finally, MoS2 nanosheets inhibited the growth of Escherichia coli in the presence of sodium nitrate. Taken together, MoS2 nanosheets triggered cellular effects as well as acellular antibacterial effects, and we provided evidence for nitrite reductase-like properties of MoS2.
1. Introduction
Bacterial infections pose a considerable burden on public health and novel approaches to overcome antimicrobial resistance are needed. Engineered nanomaterials can be used to deliver conventional antibiotics in a targeted manner or could be deployed as antibacterial agents per se to combat bacterial infections.1 However, safety concerns may prevent clinical translation of novel materials, and careful assessment of their potential adverse effects on human health is required.2Neutrophils, the most abundant white blood cells, play a key role in antimicrobial defense.3 Neutrophils can engulf bacteria leading to intracellular (oxidative and/or proteolytic) killing of pathogens, but these cells also emit neutrophil extracellular traps (NETs) resulting in the extracellular killing of bacteria and fungi.4 NETs are web-like structure consisting of a scaffold of decondensed chromatin decorated with antimicrobial granule proteins including neutrophil elastase (NE) and myeloperoxidase (MPO).5 Previous work has suggested that NETs are selectively induced by large microorganisms (e.g., fungi) thereby preventing the dissemination of the offending pathogen.6 Interestingly, we previously found that large graphene oxide (GO) sheets were more effective in provoking NETs in neutrophils than small GO sheets.7 Moreover, small and large GO sheets were both susceptible to MPO-dependent degradation in NETs.8 However, the interactions of other 2D materials with neutrophils with respect to NETs have not been investigated previously. On the other hand, several studies have suggested that graphene and related materials, as well as other emerging 2D materials such as transition metal dichalcogenides (TMDs) and transition metal carbides and nitrides (MXenes), have antibacterial properties. For instance, Tu et al.9 provided experimental and theoretical evidence that GO sheets could extract lipids from bacterial membranes, suggesting a “physical” antibacterial effect. Moreover, Li et al.10 reported that hydrated GO sheets with a high density of carbon radicals were capable of killing drug-resistant bacteria. In a more recent study, graphdiyne oxide was found to kill Escherichia coli and Staphylococcus aureus through a reactive oxygen species (ROS) dependent mechanism.11 Studies published in this journal have shown that 2D nanosheets of MoS2 exert antibacterial effects, either through lipid extraction or through a ROS-dependent pathway.12,13 Ti-based MXenes also kill bacteria, possibly through a direct effect on bacterial membranes.14 However, the observed effects (frequently using very high doses of the materials, up to 500 μg mL−1 or more) should be considered in light of the possible toxicity of the material towards host cells.
Nanomaterials, including 2D materials, may display enzyme mimetic properties including peroxidase-like or catalase-like effects.15 For instance, MoS2 has been shown to exhibit activities of four major cellular antioxidant enzymes namely superoxide dismutase, catalase, peroxidase, and glutathione peroxidase.16 Interestingly, MoS2 nanosheets were found to enhance CO2 fixation in cyanobacteria through the reprogramming of metabolic pathways and the underlying mechanism was linked to the intrinsic peroxidase-like activity of MoS2, whereas the soluble molybdate salt showed no such effects.17 However, investigations concerning the enzyme mimetic properties of 2D materials are mostly focused on the catalytic activity for ROS generation and on subsequent biological effects, whereas the capacity for reactive nitrogen species (RNS) production remains to be explored. Here, we set out to investigate the antibacterial effects of MoS2. Using HL-60 cells and primary human neutrophils, we found that MoS2 nanosheets and Na2MoO4 triggered NETs through a nitric oxide/peroxynitrite-dependent pathway. In addition, we showed that the intrinsic catalytic properties of MoS2 nanosheets can be expanded to include nitrite reductase-like effects, as evidenced by antibacterial effects of MoS2 towards E. coli in the presence of NaNO2. The acellular generation of NO was confirmed by electron paramagnetic resonance (EPR) analysis. Our study sheds light on the interactions of MoS2 with neutrophils and bacteria, and paves the way for developing novel antimicrobial agents using MoS2.
2. Results
2.1. MoS2 sheets trigger NET release in differentiated HL-60 cells
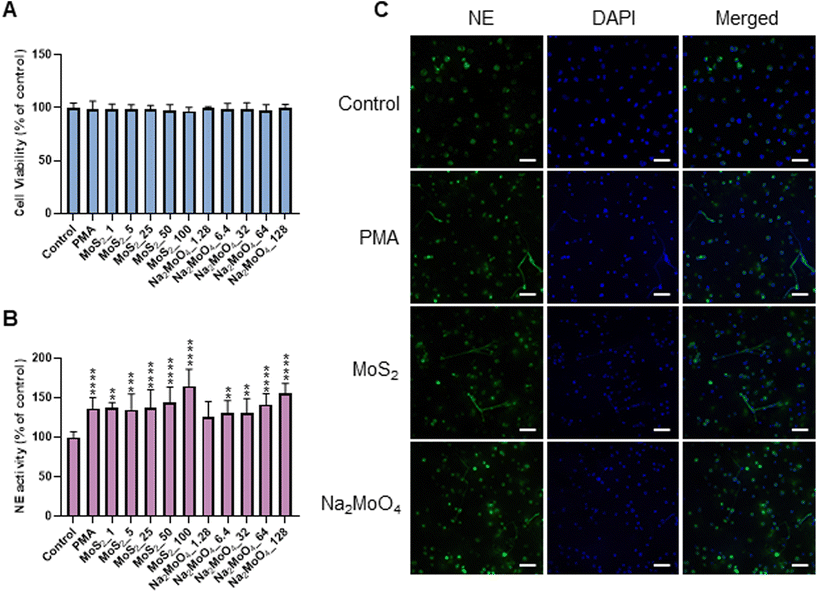
Few-layered MoS2 nanosheets were synthesized as described previously.18 The lateral size as determined by TEM analysis was 136.00 ± 79.28 nm (Fig. S1A and B†). Importantly, the endotoxin content of the 2D sheets at 25 and 50 μg mL−1 was zero or very low (well below the FDA-mandated limit for medical devices)19 (Fig. S1C†). To investigate the interaction of MoS2 sheets with neutrophils, we first tested the ability of MoS2 to trigger NETs in differentiated neutrophil-like HL-60 cells, a robust model of human neutrophils.20 Neither MoS2 nor the soluble molybdenum salt Na2MoO4 elicited any cytotoxicity (Fig. 1A). However, a significant increase of extracellular NE activity was observed after exposure to MoS2, indicative of the release of NETs.21 In fact, at the highest dose (100 μg mL−1), MoS2 was an even stronger trigger of NETs than the positive control, phorbol 12-myristate 13-acetate (PMA) (Fig. 1B). The molybdate salt also triggered a significant and dose-dependent increase of NE activity compared to untreated controls (Fig. 1B). To verify the presence of NETs, we performed immunofluorescence analysis.21 As shown in Fig. 1C, PMA, MoS2, and Na2MoO4 triggered the formation of NETs revealed by the presence of extracellular fibers decorated with NE (green). Nuclei were counterstained with DAPI (blue). In the untreated control cells, intracellular NE staining was observed. Thus, using neutrophil-like HL-60 cells as a model, MoS2 nanosheets and soluble molybdenum salt triggered NETs without causing cell death.2.2. NET release induced by MoS2 nanosheets is NO-dependent
The phorbol ester PMA activates NADPH oxidase to trigger NETs.22 Furthermore, NO, by augmenting free radical formation in neutrophils, also promotes the release of NETs.23 We asked if NO is involved in the release of NETs in the current model. To this end, the NO donor, SNAP, and the NO scavenger, cPTIO, were deployed. Cellular NO production was determined by the DAF-FM-DA assay.24 As expected, SNAP triggered significant NO production (more than 5-fold) compared to untreated control, and cPTIO significantly blocked NO generation (Fig. S2A†). Moreover, MoS2 nanosheets triggered dose-dependent NO production, which was especially evident at the highest doses (50 and 100 μg mL−1). Moreover, Na2MoO4 showed a similar trend in terms of NO production (Fig. S2A†). Notably, scavenging of NO with cPTIO led to a significant inhibition of NE activity in both MoS2 and Na2MoO4 exposed cells, indicating that NO played a role in NET release (Fig. S2B†). Cellular NO and superoxide react rapidly to form peroxynitrite, and a previous study has shown that scavenging of peroxynitrite inhibited NO-induced NET formation.25 We thus investigated the role of peroxynitrite in our model. First, we asked whether the MoS2 nanosheets themselves were susceptible to peroxynitrite. To this end, MoS2 nanosheets were incubated with SIN-1, a peroxynitrite donor previously shown to elicit the degradation of GO.24 The dispersion of MoS2 became completely translucent at 3 h (Fig. S3A†), suggestive of biotransformation, and further analysis revealed a red-shift of the Raman spectra (Fig. S3B†). Previous work has attributed this shift to doping,26 and we thus interpret this as a sign of nitrogen doping of MoS2. However, the two Raman modes E12g and A1g, known to exhibit thickness dependence,27 were not affected (Fig. S3B†). Next, we assessed peroxynitrite production in neutrophil-like HL-60 cells. To this end, the DAX-J2™ PON Green probe was used to detect peroxynitrite, and DPI and L-NAME, pharmacological inhibitors of NADPH oxidase and NOS, respectively, were applied (Fig. 2A). As shown in Fig. 2B, MoS2 nanosheets as well as Na2MoO4 triggered peroxynitrite production which was blocked by DPI and L-NAME. PMA and SNAP were included as positive controls. We then applied the extracellular NE activity-based assay to test whether peroxynitrite production was required for NET release. As shown in Fig. 2C, DPI and L-NAME were able to inhibit NE activity significantly in cells exposed to MoS2 as well as Na2MoO4. These effects were further confirmed by immunofluorescence staining of NE, as shown in Fig. S4.† Collectively, our results demonstrated that NO is involved in the release of NETs in cells exposed to MoS2. It is conceivable that MoS2 nanosheets undergo biotransformation in contact with cells leading to the release of Mo ions which, in turn, contributes to the NET release.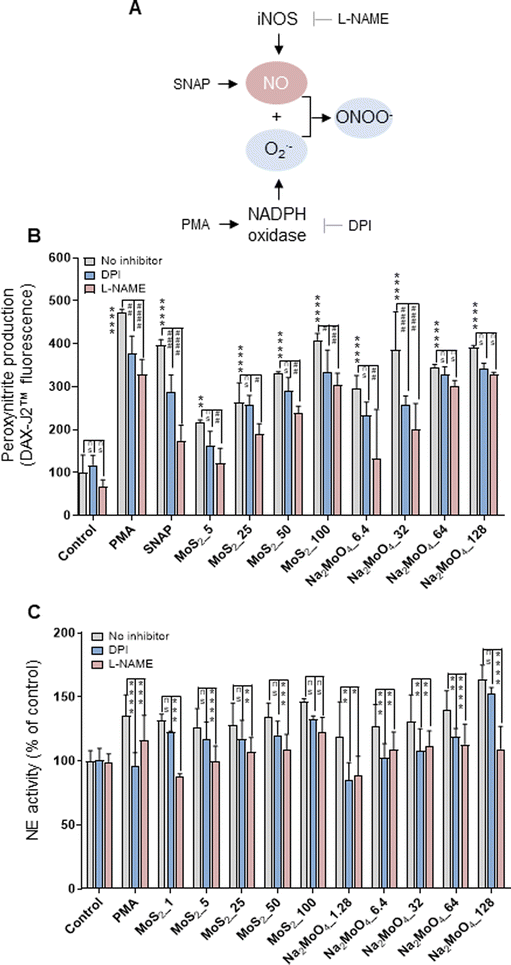 | ||
| Fig. 2 Role of peroxynitrite for NET release. (A) Cartoon showing the role of iNOS and the NADPH oxidase for the production of peroxynitrite. (B) Quantification of peroxynitrite production using the DAX-J2™ PON Green probe. The differentiated HL-60 cells were preincubated with DPI (10 μM) and L-NAME (15 mM) for 30 min prior to the exposure to PMA (50 nM), SNAP (500 μM), MoS2, and Na2MoO4 at the indicated concentrations for 3 h. Peroxynitrite production was observed in cells exposed to MoS2 and Na2MoO4, which was blocked by DPI and L-NAME. Two-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, #p < 0.05; ##p < 0.01, ###p < 0.001, ####p < 0.0001; ns = no significance). (C) Role of peroxynitrite for NET release quantified by extracellular NE activity. DPI (10 μM) and L-NAME (15 mM) were used to inhibit NADPH oxidase and NOS, respectively. Data shown as mean values ± S.D. (n = 3). Two-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = no significance). Refer to Fig. S2† for the effect of the NO scavenger cPTIO on NET production, and Fig. S4† for the visualization of NETs by immunofluorescence. | ||
2.3. MoS2 nanosheets trigger NETs in primary neutrophils
To verify our findings using HL-60 cells, we investigated whether MoS2 triggered NET release in primary human neutrophils. In line with the observations in differentiated HL-60 cells, MoS2 and Na2MoO4 did not cause significant cytotoxicity in primary cells (Fig. S5A†), while a dose-dependent NET release was found as evidenced by a significant increase of extracellular NE activity (Fig. S5B†). The formation of NETs was confirmed by immunofluorescence staining which revealed typical extracellular staining of NE (green) in the PMA, MoS2, and Na2MoO4 exposed samples (Fig. 3). Furthermore, the MoS2 nanosheets appeared to attach to the NETs (Fig. S6A†). To verify this, we performed SEM analysis of primary neutrophils exposed to PMA and MoS2. We obtained clear evidence of web-like NETs, and it appeared that MoS2 sheets were attached to the chromatin fibers (Fig. S6B†). Our results thus demonstrated that MoS2 nanosheets as well as Na2MoO4 triggered NET release in primary human neutrophils in the absence of cell death.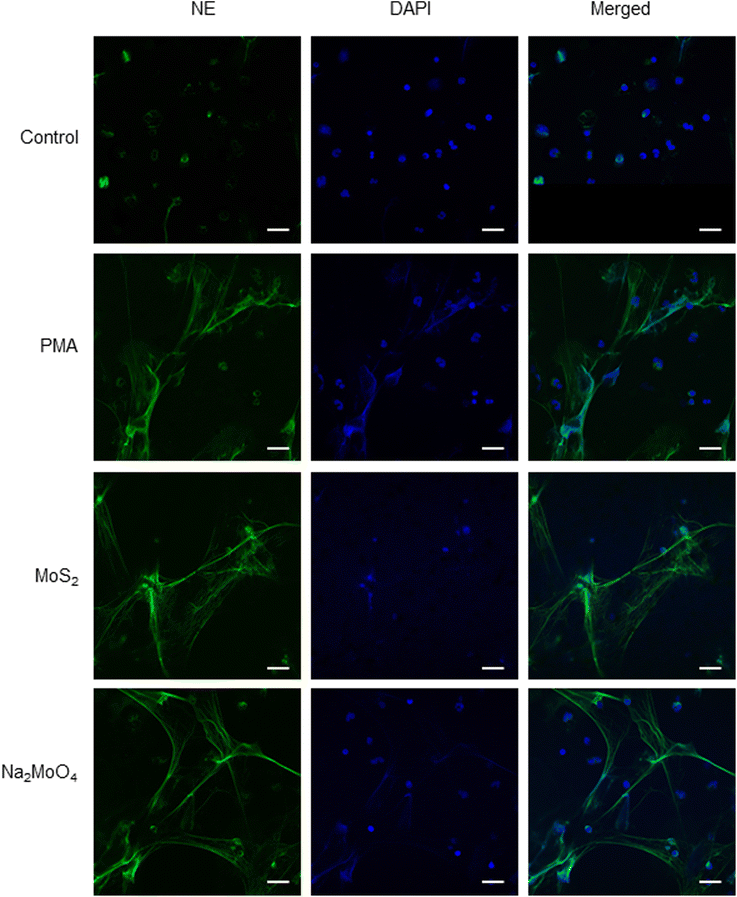 | ||
| Fig. 3 Endotoxin-free MoS2 nanosheets trigger NETs in primary human neutrophils. Visualization of NETs by immunofluorescence staining of NE. MoS2 (50 μg mL−1) and Na2MoO4 (64 μg mL−1) triggered vigorous release of NETs in freshly isolated human neutrophils at 3 h. PMA (25 nM) was used as positive control. Scale bars: 20 μm. Refer to Fig. S5† for data on cell viability following exposure to MoS2 and Na2MoO4 and Fig. S6† for further evidence of NETs in primary cells using SEM. | ||
2.4. MoS2 nanosheets induce acellular bacterial killing
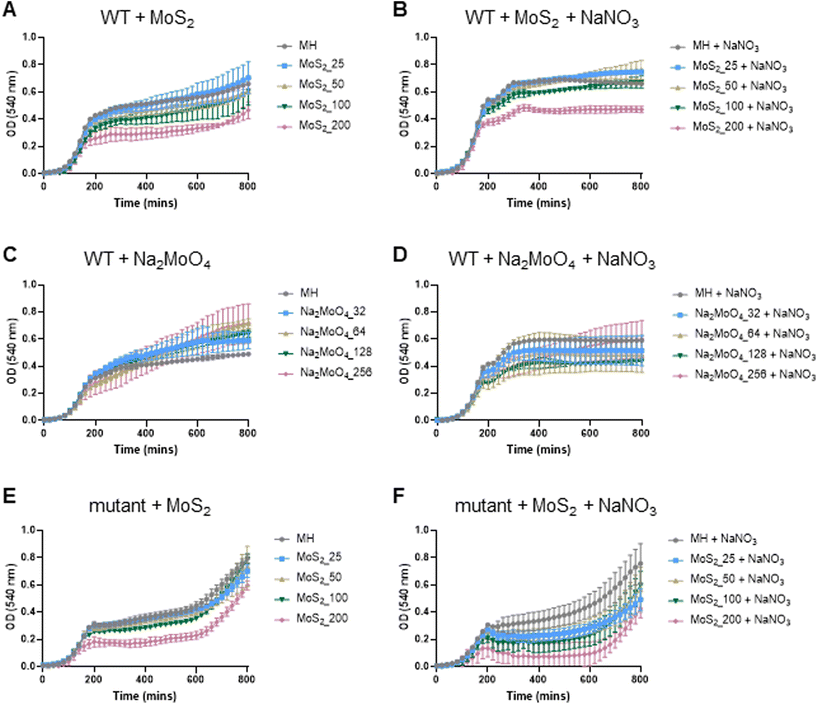
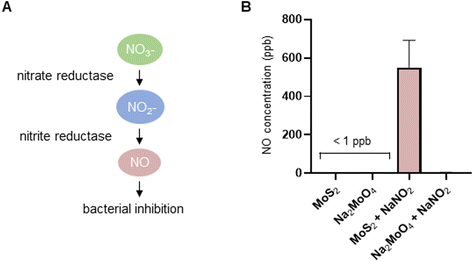
The experiments presented above suggested that MoS2 may promote antibacterial effects through its effects on neutrophils, but any direct effects of MoS2 on bacteria are difficult to evaluate using these models. To address whether MoS2 nanosheets possess intrinsic antibacterial properties, and whether this could be linked to enzyme mimetic effects, we performed a series of acellular experiments. Specifically, we asked whether MoS2 mimicked the properties of nitrite reductases (enzymes that catalyze the reduction of nitrite) with subsequent antibacterial effects. To test this, we explored the inhibition of bacterial growth by MoS2 nanosheets using the E. coli RK 4353 wild-type strain versus the mutant strain lacking nitrate reductases.28 We first confirmed that the wild-type strain, but not the mutant, was able to utilize nitrate (NaNO3) to promote its growth (Fig. S7†). We then determined the growth of E. coli in the presence or absence of MoS2 or Na2MoO4 with or without the addition of NaNO3 (Fig. 4A–D). The results revealed that MoS2 nanosheets inhibited the log-phase growth of wild-type E. coli in a dose-dependent manner (Fig. 4A), and the addition of NaNO3 further enhanced the suppression of bacterial growth, particularly at the highest dose of MoS2 (200 μg mL−1) (Fig. 4B). In comparison, exposure to Na2MoO4 at equivalent Mo concentrations did not impact on bacterial growth (Fig. 4C), nor did the addition of NaNO3 elicit obvious antibacterial effects (Fig. 4D). Interestingly, synergistic antibacterial effects were confirmed by supplementation with the NO donor, DETA NONOate (Fig. S8†). Furthermore, unlike the observations in the wild-type strain, MoS2 nanosheets inhibited the growth of mutant E. coli only at the highest dose (200 μg mL−1) (Fig. 4E), and the addition of NaNO3 did not promote the antibacterial effects elicited by the MoS2 nanosheets (Fig. 4F). However, the observed effects were modest and should be taken as indicative rather than definitive. Nevertheless, the differences between wild-type and mutant in the presence of NaNO3, as well as the differences between MoS2 and Na2MoO4, suggested that MoS2 nanosheets might mimic nitrite reductases to produce NO, which consequently may account for the antibacterial effects. To verify our hypothesis that MoS2 nanosheets catalyzed the production of NO from NaNO2 (Fig. 5A) we investigated acellular NO production using two different approaches. First, incubation of MoS2 nanosheets with nitrite (NaNO2) generated detectable amount of NO (Fig. 5B), as determined by chemiluminescence.29 In contrast, soluble Na2MoO4 did not produce NO with NaNO2, indicating that the effects were nanomaterial/nanosheet specific. The generation of NO radicals was further confirmed by EPR analysis using the nitronyl nitroxide PTIO as the spin trap reagent. The reaction of PTIO with NO led to the typical EPR spectra of immino nitroxide and nitrogen dioxide (Fig. S9A and B†). Our results demonstrated that MoS2 nanosheets mixed with NaNO2 generated NO radicals in a dose (Fig. 6A and C) and time dependent manner (Fig. 6B and D). Cyclic voltammetry (CV) can be used to acquire information about the redox behavior of nanomaterials.30,31 To provide further evidence for the nanozyme-like properties of 2D MoS2 nanosheets, we performed CV to ascertain electrochemical nitrite reduction. We found that the cathodic current was enhanced from 0.02 V (Fig. 6E) to 0.17 V (Fig. 6F) in the presence of nitrite (NaNO2) (and refer to Fig. S10† for the individual CV scans). In summary, the 2D MoS2 nanosheets seemed to possess enzyme mimetic properties.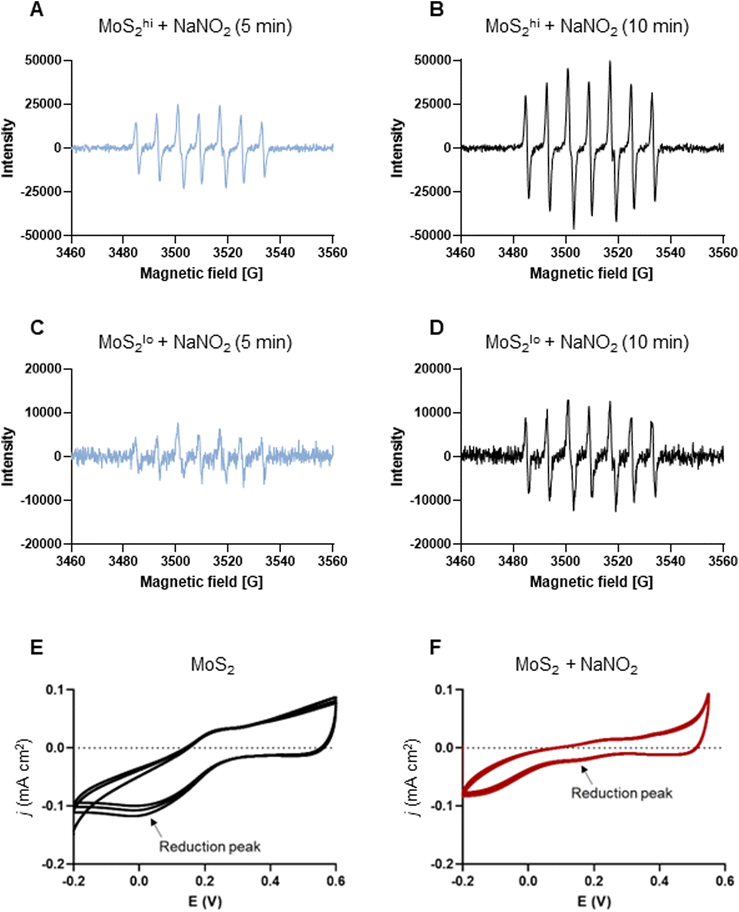 | ||
| Fig. 6 MoS2 nanosheets produce NO radicals: evidence for nanozyme properties. (A–D) EPR analysis of NO radicals (and refer to Fig. S9†). Two doses of MoS2 (high and low) (1 mg mL−1 and 200 μg mL−1) were mixed with NaNO2 at the dose of 10 mM. The spin trap PTIO (100 mM) was added immediately prior to the analysis, and EPR spectra were captured at 5 min and 10 min. (A and B) MoS2 nanosheets (1 mg mL−1) mixed with sodium nitrite (NaNO2) (10 mM) generated NO radicals captured by the spin trap, PTIO (100 mM) at 5 min (A) and 10 min (B). (C and D) MoS2 (200 μg mL−1) mixed with NaNO2 (10 mM) generated NO radicals captured by PTIO at 5 min (C) and 10 min (D) albeit not to the same extent as high-dose MoS2. Redox transition of MoS2 nanosheets in the absence (E) or presence (F) of nitrite as evidenced by cyclic voltammetry. Upon addition of nitrite, the cathodic current was enhanced from 0.02 V to 0.17 V (arrows), as a result of the redox process. | ||
3. Discussion
In the present study, we investigated the potential antibacterial properties of MoS2 nanosheets and the underlying mechanisms using cellular and acellular systems. We demonstrated for the first time that MoS2 triggered the release of anti-bacterial NETs, and this was shown to occur through an NO/peroxynitrite-dependent pathway. Importantly, MoS2 triggered the release of NETs in neutrophil-differentiated HL-60 cells as well as in primary human neutrophils at doses that were non-cytotoxic towards these cell types. We also found that the soluble molybdenum salt (Na2MoO4) triggered NETs. This is consistent with a recent study showing that sodium molybdate induced extracellular trap formation in heterophils in the absence of cell death.32 In addition, a direct antibacterial effect of MoS2 nanosheets was demonstrated in the present study, and evidence was provided for NO production, attributed to the enzyme mimetic properties of MoS2. Indeed, EPR analysis as well as electrochemical analysis supported the conclusion that 2D MoS2 nanosheets display enzyme-like activity, and these effects were not seen for the soluble Mo salt. Non-enzymatic mechanisms such as a direct physical damage of bacterial membranes, or a combination of physical effects and metabolic reprogramming,33 cannot be ruled out, as the effects on bacterial growth were observed in wild-type and mutant bacteria in the absence of NaNO3. However, in the presence of NaNO3, the impact of MoS2 on E. coli was more pronounced, and the results point to a role of NO in bacterial killing. The present study thus points to a novel antibacterial mechanism which exploits the propensity of nitrate-reducing bacteria to generate nitrite, which is then transformed by MoS2 to NO. Taken together, these results expand the repertoire of so-called nanozyme (enzyme mimetic) activities of MoS2.34 Thus, while previous studies emphasized peroxidase-like, catalase-like, and superoxide dismutase-like properties, our study is the first to provide evidence of a nitrite reductase-like effect. However, it is important to recognize that a true “nanozyme” should have effects similar to enzymes, i.e., efficient catalytic activity, and a specific mechanism of action.35 Therefore, we prefer the term “nanozyme-like”.NETs are tasked with the clearance of invading pathogens including bacteria, and the involvement of NADPH oxidase-dependent and independent pathways are well documented.22,36 Furthermore, RNS (i.e., NO and peroxynitrite) have also been shown to enhance the release of NETs.23,25 In our previous studies, we could show that GO sheets triggered NETs in a size-dependent manner inasmuch as small GO sheets triggered canonical NADPH oxidase-dependent release of NETs while large GO sheets triggered NETs in an NADPH oxidase-independent manner.7 It is pertinent to note that carbon-based nanomaterials are also susceptible to degradation through NADPH oxidase and/or iNOS-driven pathways,8,21,24,37,38 thus suggesting reciprocal interactions between nanomaterials and the immune system.3 However, in the present study, the release of NETs is shown to be Mo-dependent, suggesting a different mechanism (in other words, not size-dependent as shown for GO).
Cellular NO generation occurs via the oxidative route, with the involvement of nitric oxide synthase (NOS), or via the nitrate-nitrite-dependent reductive route.39 NOS enzymes produce NO from arginine, and neutrophils as well as classically activated macrophages24 express inducible NOS (iNOS), which accounts for the NO production observed in cells encountering MoS2 nanosheets. The fact that the NO scavenger cPTIO, as well as a NADPH oxidase inhibitor (DPI) and a NOS inhibitor (L-NAME) prevented NET formation in our model supports the conclusion that MoS2 nanosheets triggered NETs through an NO/peroxynitrite-driven pathway. It is worth noting that early work implicated NO in NET release insofar as L-NAME was found to reduce NET release in mouse neutrophils.40 Moreover, the NO donor SNAP was shown to trigger NETs.23,25 However, NADPH oxidase activity was required to release NETs upon stimulation with NO, thus implying a role for peroxynitrite.25 We confirmed here that SNAP triggered release of NETs in neutrophils. Nevertheless, the involvement of ROS in NET formation cannot be dismissed.22 Indeed, this may provide an explanation for the observation that SNAP triggered the highest level of NO, but prompted less NETs as compared to the MoS2 nanosheets. Thus, while NET release may be regarded as an NO-dependent process, its metabolite, peroxynitrite appears to be the key factor in NET release from cells exposed to MoS2.
NETs were initially shown to play a pivotal role in host defense by immobilizing and destroying pathogens (bacteria as well as fungi).5,41 However, it is pertinent to note that excessive or inadvertent NET formation, or a lack of clearance of NETs by macrophages or other phagocytic cells,42 is a risk factor for autoimmune disease and thrombosis and may lead to vascular occlusion.43 In other words, while NETs display antimicrobial properties, they may be viewed as a double-edged sword of innate immunity.44 Moreover, some authors have coupled the release of NETs with a necrotic cell death dubbed NETosis.45 However, in the present study, the NET-releasing cells remained viable. Further studies, including studies in relevant animal models, are required to understand the consequences of MoS2-driven NET release.
Interestingly, all members of the molybdenum (Mo)-dependent enzyme family have been shown to reduce nitrite to NO.46 Mao et al.47 showed in a very recent study that Na2MoO4 triggered NO production in ovarian cancer cells, and the authors found that the simultaneous silencing of SUOX, AOX1, and mARC1 in the SKOV3 cell line lowered the Na2MoO4-induced NO levels, implying a role for these Mo-dependent enzymes, while the silencing of XDH or mARC2 had no impact on NO production. Thus, bioavailable Mo could yield NO through more than one pathway. Cao et al.48 reported that MoS2 may undergo biotransformation (oxidation) in vivo, donating its Mo towards the synthesis of Mo-dependent enzymes in the liver. However, NO production was not investigated in the latter study. The present data support the view that MoS2-triggered NO generation in neutrophils occurs through an iNOS-dependent pathway. However, it is necessary to consider the reductive pathway of NO production, and “the versatile redox chemistry of molybdenum”49 to understand the acellular MoS2-triggered production of NO (in the presence of nitrite).
Living organisms use nitrite for a variety of different purposes.50 One of us previously demonstrated that acidification of nitrite-rich urine resulted in the killing of nitrate-reducing bacteria.28 Here, we explored the enzyme mimetic activity of MoS2 nanosheets, specifically whether the nanosheets could function as nitrite reductases, and the subsequent impact on bacterial growth. Taking advantage of wild-type E. coli and a mutant strain lacking nitrate reductases, we could show that MoS2 nanosheets elicited more pronounced bacterial inhibition in wild-type bacteria in the presence of sodium nitrate (NaNO3). Moreover, EPR analysis provided evidence for the presence of NO radicals in the system. Importantly, soluble Na2MoO4 did not evoke similar effects, which further suggested that the enzyme-like activity evidenced here is a 2D material-specific property. MoS2 was shown previously to act as a catalyst for denitrification, thus facilitating the electrochemical reduction of nitrate/nitrite to ammonia.51 Previous work has also shown that the catalase-like nanozyme activities of MoS2 are more pronounced under acidic conditions.16 The present acellular studies were performed at neutral pH. Further studies are needed to determine whether the nitrite reductase-like properties of MoS2 nanosheets are pH-dependent. As noted above, acidification of urine is known to promote the killing of nitrate-reducing bacteria likely due to a conversion of nitrite to bactericidal nitrogen oxides.28 It may also be important to consider the impact of pH on the nanosheets themselves. Indeed, Wang et al.52 demonstrated in a comprehensive study that MoS2 may undergo oxidative dissolution with kinetics that depend on pH.
The present study is a proof-of-concept study. Nevertheless, we foresee several potential applications of MoS2 nanosheets including in the clinical setting, e.g., coating of catheters or medical implants using MoS2 nanosheets to combat infections and prevent the formation of biofilms. Moreover, nanozyme-like MoS2 nanosheets could potentially be incorporated in wound dressings, perhaps as hydrogels, as shown recently in an elegant study by Sang et al.53 Beyond such biomedical applications, MoS2 could also be integrated into water filtration membranes used in environmental remediation processes.54 To our knowledge, the antibacterial properties of MoS2 nanosheets have not been fully exploited in this setting.
4. Conclusions
Our study has provided evidence that MoS2 nanosheets evoke NO-dependent antibacterial effects. These effects occurred indirectly through effects on neutrophils, leading to the release of NETs, and directly by virtue of enzyme mimetic properties. Hence, in the cellular system, NO/peroxynitrite contributed to the release of NETs, which are known to kill bacteria (and fungi). This was also observed for sodium molybdate. In the acellular system, NO generated via the nitrate-nitrite-dependent reductive route contributed to the antibacterial effects of MoS2. The present study is thus the first to suggest the possibility of nitrite reductase-mimicking properties of MoS2.5. Experimental section
5.1. Synthesis and characterization of MoS2
The 2D MoS2 nanosheets were synthesized according to a previously established protocol.18 Briefly, bulk molybdenum disulfide (Sigma-Aldrich) was used as the precursor material and glycine (Sigma-Aldrich) was used as the exfoliating agent. The mechanochemical process was conducted in a ball-milling machine (Retsch pm100) at room temperature and air atmosphere for 15 min at the speed of 250 rpm. After the milling treatment, the resulting solid mixtures were dispersed in water and dialyzed to remove the glycine. Dry powder samples were obtained after lyophilization at −80 °C at a pressure of 0.005 bar. The powders were stored at room temperature and dispersions were obtained by dispersing in water, followed by water bath sonication (10 s cycles for 2 min). TEM analyses were performed on stable dispersions of MoS2 diluted as necessary and dip-casted on Lacey copper grids (3.00 mm, 200 mesh), coated with carbon film, and dried under vacuum. Samples were investigated using a HR-TEM JEOL 2100 at an accelerating voltage of 100 kV. Further characterization results for as-produced samples are reported in González et al.18 and are not replicated here. For Raman spectroscopy, see below. Sodium molybdate (Na2MoO4) (CAS number: 7631-95-0) was obtained from Sigma. The stock solution was prepared at a concentration of 1.280 mg mL−1 (i.e., the equivalent amount of Mo present in 1 mg mL−1 MoS2) in endotoxin-free ultrapure water.5.2. Raman spectroscopy
For Raman spectroscopy,55 MoS2 nanosheets exposed or not to the peroxynitrite donor, 3-morpholinosydnonimine hydrochloride (SIN-1) (Sigma-Aldrich) were deposited by drop-casting on SiO2/Si substrate, blow dried by N2 gas, washed three times in dH2O, followed by Raman analysis which was carried using the Witec Alpha 300 RAS instrument. The 532 nm laser optimized to 3 mW to minimize heating was used to acquire data. Data shown are the average of at least 5 spectra per sample.5.3. Limulus amebocyte lysate assay
Endotoxin content of the MoS2 samples was evaluated using the Pierce™ Chromogenic Endotoxin Quant Kit (ThermoFisher Scientific). Briefly, MoS2 nanosheets (25 and 50 μg mL−1) were mixed with LAL solution and incubated at 37 °C for 10 min. The absorbance values were captured at 405 nm using a Tecan Infinite® F200 plate reader (Stockholm, Sweden). MoS2 (50 μg mL−1) spiked with lipopolysaccharides (LPS) (0.5 EU mL−1) was included as an interference test (not shown).5.4. HL-60 cell differentiation and culture
The human acute promyelocytic leukemia cell line HL-60 (ATCC-CCL-240) was maintained in phenol red-free RPMI-1640 medium supplemented with 2 mM L-glutamine, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, and 10% heat-inactivated fetal bovine serum (FBS) (Sigma). In order to achieve neutrophil-like differentiation,20 the cells were seeded at 0.5 × 106 cells per mL in cell medium supplemented with 1.25% DMSO for 5 days. The medium was changed after 3 days.5.5. Human neutrophil isolation and culture
Neutrophils were isolated from buffy coats obtained from healthy adult blood donors (Karolinska University Hospital, Stockholm, Sweden) as previously described.8 In brief, neutrophils were separated by density gradient centrifugation with Lymphoprep™ (Stemcell™ Technologies, Sweden). The neutrophil layer was then subjected to sedimentation in 5% dextran solution, followed by hypotonic lysis of residual erythrocytes using deionized water. Isolated neutrophils were maintained in phenol red-free RPMI-1640 culture medium (Sigma) supplemented with 2 mM L-glutamine, 100 U mL−1 penicillin, and 100 mg mL−1 streptomycin without fetal bovine serum.5.6. Cytotoxicity assessment
Cell viability assessment following MoS2 and Na2MoO4 exposure was evaluated based on the ATP content by using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega). Cells were seeded in 96-well plates (1 × 106 cells per mL) in phenol red-free RPMI-1640 medium supplemented with L-glutamine, penicillin, and streptomycin. Cells were exposed for 3 h to MoS2 nanosheets (1, 5, 25, 50, and 100 μg mL−1) and to the equivalent concentration of Na2MoO4 (1.28, 6.4, 32, 64, and 128 μg mL−1). PMA (Sigma-Aldrich) was used as a positive control (50 nM in differentiated HL-60 cells20 and 25 nM in primary neutrophils42). Then, CellTiter-Glo® reagent was added, and plates were shaken for 2 min and incubated at room temperature for 10 min. Luminescence values were recorded using a Tecan Infinite® F200 plate reader.5.7. Neutrophil elastase activity
NE activity in the cell culture supernatants was analyzed as described previously.7 Briefly, cells were seeded in 96-well plates at a density of 1 × 106 cells per mL. After 3 h of exposure to MoS2 (1, 5, 25, 50 and 100 μg mL−1) and Na2MoO4 (1.28, 6.4, 32, 64, and 128 μg mL−1), the supernatants were collected after intensive pipetting and centrifuged at 1600 rpm for 5 min. PMA was used as a positive control for the induction of NETs. The supernatants were incubated with 5 μg mL−1 of DNase I (Sigma-Aldrich) for 15 min at room temperature to digest the DNA,56 followed by incubation with the elastase substrate N-(methoxysuccinyl)-Ala-Ala-Pro-Val 4-nitroanilide (200 μM) (Sigma-Aldrich) in the assay buffer (Tris 0.1 M, NaCl 0.5 M, pH 8). After incubation for 3 h at room temperature, the absorbance at 405 nm was recorded using a Tecan Infinite® F200 plate reader. The NE activity was normalized to untreated controls. To investigate the role of NO and peroxynitrite, the NO donor, S-nitroso-N-acetylpenicillamine (SNAP, 500 μM) (Sigma-Aldrich) and the NO scavenger, 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO, 250 μM) were used, and diphenyleneiodonium chloride (DPI, 10 μM), and Nω-nitro-L-arginine methyl ester hydrochloride (L-NAME, 15 mM) (all from Sigma-Aldrich) were used to block NADPH oxidase and iNOS, respectively. For these experiments, cells were pre-incubated with cPTIO, DPI and L-NAME for 30 min prior to the exposure to MoS2 nanosheets, Na2MoO4, and positive controls. The supernatants were collected, and NE activity was determined as described above.5.8. Confocal microscopy of NETs
To visualize NETs, we performed confocal microscopy as described.20 Sterilized coverslips were coated with poly-L-lysine (Sigma-Aldrich) for 20 min, washed three times with PBS, and dried at room temperature. Differentiated HL-60 cells or primary neutrophils were seeded on the coverslips at a density of 1 × 106 cells per mL for 1 h. Then, HL-60 cells were exposed to PMA (50 nM), MoS2 (5 and 50 μg mL−1) and Na2MoO4 (6.4 and 64 μg mL−1) for 3 h. For DPI and L-NAME groups, the cells were pre-incubated with DPI (10 μM) and L-NAME (15 mM) for 30 min. For primary neutrophils, cells were exposed to the PMA (25 nM), MoS2 (50 μg mL−1) and Na2MoO4 (64 μg mL−1) for 3 h. After the exposure, the slides were fixed with 4% paraformaldehyde solution for 30 min at RT, followed by blocking for 30 min in 2% BSA–PBS solution. The cells were then stained using a mouse anti-human NE antibody (sc-53388, Santa Cruz Biotechnology) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in 2% BSA–PBS) at 4 °C overnight. The secondary antibody staining was performed using the Alexa-488 goat anti-mouse antibody (1
100 in 2% BSA–PBS) at 4 °C overnight. The secondary antibody staining was performed using the Alexa-488 goat anti-mouse antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in 2% BSA–PBS, ThermoFisher Scientific) for 1 h at room temperature. The slides were then washed three times with PBS and mounted with ProLong™ Gold Antifade Mountant with DAPI (ThermoFisher Scientific). The confocal images were captured using a Zeiss LSM880 confocal microscopy (ZEISS).
500 in 2% BSA–PBS, ThermoFisher Scientific) for 1 h at room temperature. The slides were then washed three times with PBS and mounted with ProLong™ Gold Antifade Mountant with DAPI (ThermoFisher Scientific). The confocal images were captured using a Zeiss LSM880 confocal microscopy (ZEISS).
5.9. Scanning electron microscopy
Primary neutrophils (1 × 106 cells per mL) were seeded in 24-well plates on poly-L-lysine coated coverslips. After 1 h, cells were exposed for 3 h to PMA (25 nM), MoS2 (50 μg mL−1), or Na2MoO4 (64 μg mL−1). SEM was then performed as described.7 Briefly, slides were fixed with 2.5% glutaraldehyde in 0.1 M PBS, rinsed in dH2O, placed in 70% ethanol for 10 min, followed by 95% ethanol for 10 min, absolute ethanol for 15 min, and, finally, acetone. Samples were dried with a critical point dryer (Balzer, CPD 010) and CO2, mounted on an aluminum stub, coated with carbon (Bal-Tec MED 010), and analyzed with an Ultra 55 field emission microscope (Zeiss) at 3 kV.5.10. NO and peroxynitrite detection
NO production in differentiated HL-60 cells was determined by the DAF-FM-DA assay (Sigma-Aldrich) as described previously.57 Cells were preincubated for 30 min with cPTIO (250 μM) to scavenge NO. After 3 h exposure to PMA (50 nM), SNAP (500 μM), MoS2 (1, 5, 25, 50 and 100 μg mL−1) and Na2MoO4 (1.28, 6.4, 32, 64, and 128 μg mL−1), cells were washed with PBS and incubated with DAF-FM-DA (10 μM) for 60 min. Then, cells were washed with PBS to remove excess probe and incubated for 30 min to allow complete de-esterification. The fluorescence signal was captured at the excitation and emission wavelengths of 495 and 515 nm using a Tecan Infinite® F200 plate reader. Peroxynitrite generation in neutrophil-like HL-60 cells was determined using the DAX-J2™ PON Green probe (AAT BioQuest, Sunnyvale, CA), as previously described.24 DPI (10 μM) and L-NAME (15 mM) were used to block the NADPH oxidase and iNOS, respectively. After 3 h of exposure to PMA, MoS2, or Na2MoO4, cells were incubated with the fluorescent probe for 2 h in the dark at 37 °C. The fluorescence signal was captured at the excitation and emission wavelengths of 490 and 530 nm using a Tecan Infinite® F200 plate reader.5.11. Bacterial growth inhibition
To assess the impact of MoS2 nanosheets versus Na2MoO4 on bacterial growth, E. coli RK 4353 wild-type (WT) and mutant strains lacking nitrate reductases were used.58 The E. coli strains were a generous gift from Jeff A. Cole, University of Birmingham, UK. The bacterial solutions were prepared by preincubation of two colonies in 4 mL Mueller-Hinton (MH) broth (Sigma-Aldrich) for 6 h to give a concentration of 108 CFU mL−1. Then, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution of bacterial solution was prepared with MoS2 and Na2MoO4 to yield the final concentrations of MoS2 (25, 50, 100, and 200 μg mL−1) and Na2MoO4 (32, 64, 128, and 256 μg mL−1). NaNO3 (10 mM) (Sigma-Aldrich) and DETA NONOate (100 μM) (Sigma-Aldrich) was supplied as indicated. The bacterial solutions for each condition were transferred to flat-bottom microwell plates (Corning) using three wells (technical replicates) per condition. Bacterial growth was measured continuously for 800 min at 37 °C and absorbance values at 540 nm was recorded every 20 min in a computerized incubator (Spectra Max 340, Molecular Devices), as previously described by us.28 MoS2 nanosheets were included as a reference and absorbance values were subtracted accordingly.
100 dilution of bacterial solution was prepared with MoS2 and Na2MoO4 to yield the final concentrations of MoS2 (25, 50, 100, and 200 μg mL−1) and Na2MoO4 (32, 64, 128, and 256 μg mL−1). NaNO3 (10 mM) (Sigma-Aldrich) and DETA NONOate (100 μM) (Sigma-Aldrich) was supplied as indicated. The bacterial solutions for each condition were transferred to flat-bottom microwell plates (Corning) using three wells (technical replicates) per condition. Bacterial growth was measured continuously for 800 min at 37 °C and absorbance values at 540 nm was recorded every 20 min in a computerized incubator (Spectra Max 340, Molecular Devices), as previously described by us.28 MoS2 nanosheets were included as a reference and absorbance values were subtracted accordingly.
5.12. Chemiluminescence analysis
Acellular NO production was determined using a chemiluminescence NO analyzer (Eco Physics, Durnten, Switzerland).29 Specifically, MoS2 (1 mg mL−1) or Na2MoO4 (1.28 mg mL−1) was mixed with sodium nitrite (NaNO2) (1 mM) (Sigma-Aldrich) in a syringe and incubated for 2 min. The air space was set as 50 mL. After 2 min incubation, 40 mL of the air was taken by another syringe and injected to the chemiluminescence NO detector for NO quantification. The NO detection limit was 1 ppb.5.13. EPR analysis for NO detection
The acellular NO radical formation was determined by EPR analysis.59 Specifically, MoS2 nanosheets (200 μg mL−1 and 1 mg mL−1) and NaNO2 (10 mM) were mixed immediately prior to the analysis. The nitronyl nitroxide, 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO) (100 mM) (Sigma-Aldrich) was added immediately to detect NO radicals.60 The EPR spectra were captured at 5 min and 10 min after adding PTIO using the Bruker spectrometer A300-10/12 (Mannheim, Germany). In addition, the EPR spectrum of PTIO alone was captured as a reference.5.14. Cyclic voltammetry
A one-compartment, three-electrode system was employed for the electrochemical nitrite reduction experiments. The electrolyte consisted of a 0.5 M Na2SO4 aqueous solution (pH = 6.8). NaNO2 was added to the electrolyte at a final concentration of 0.1 M. Electrochemical measurements were conducted using a commercial electrochemical workstation (CHI-760E, Shanghai Chenhua Instrument Co., Ltd, China). Pt wire and a saturated calomel electrode were utilized as counter and reference electrodes, respectively. The working electrode was designed using the MoS2 sample coated on the fluorine-doped tin oxide conductive glass surface. The measurements were performed by scanning from the resting potential at a scanning rate of 2 mV s−1. The CV scans were conducted three times to examine the repeatability of the electrochemical properties (refer to Fig. S10† for individual scans).5.15. Statistical analysis
Three independent experiments were performed using the HL-60 cell line and experiments with primary neutrophils were performed using cells isolated from three individual donors. The results shown are the mean values ± S.D. and the statistical analysis was performed using Student's t-test and two-way ANOVA (GraphPad, Prism 8.0.2). Statistically significant differences were considered when p < 0.05.Author contributions
G. P. performed experiments, analyzed data, and drafted the paper; V. G. performed experiments; E. V. supervised the work, and analyzed data; J. O. L. supervised the work, and analyzed data; B. F. coordinated the study, supervised the work, analyzed data, and edited the paper, and all co-authors approved the final version of the paper.Conflicts of interest
The authors declare no competing interests.Acknowledgements
Supported by the European Commission through the Graphene Flagship (grant agreement no. 881603) (B. F., E. V.). EPR and CV studies were supported by the Fundamental Research Funds for the Central Universities (G. P.). We thank Lars Haag, Electron Microscopy Core Facility, Karolinska Institutet, for assistance with SEM, Illia Dobryden, RISE Research Institutes of Sweden, for assistance with Raman, and Carina Nihlen, Department of Physiology and Pharmacology, Karolinska Institutet, for assistance with the bacterial growth inhibition studies.References
- J. M. V. Makabenta, A. Nabawy, C. H. Li, S. Schmidt-Malan, R. Patel and V. M. Rotello, Nat. Rev. Microbiol., 2021, 19, 23–36 CrossRef CAS PubMed.
- B. Fadeel, L. Farcal, B. Hardy, S. Vázquez-Campos, D. Hristozov, A. Marcomini, I. Lynch, E. Valsami-Jones, H. Alenius and K. Savolainen, Nat. Nanotechnol., 2018, 13, 537–543 CrossRef CAS PubMed.
- G. Peng and B. Fadeel, Adv. Drug Delivery Rev., 2022, 188, 114422 CrossRef CAS PubMed.
- V. Papayannopoulos, Nat. Rev. Immunol., 2018, 18, 134–147 CrossRef CAS PubMed.
- V. Brinkmann, U. Reichard, C. Goosmann, B. Fauler, Y. Uhlemann, D. S. Weiss, Y. Weinrauch and A. Zychlinsky, Science, 2004, 303, 1532–1535 CrossRef CAS PubMed.
- N. Branzk, A. Lubojemska, S. E. Hardison, Q. Wang, M. G. Gutierrez, G. D. Brown and V. Papayannopoulos, Nat. Immunol., 2014, 15, 1017–1025 CrossRef CAS PubMed.
- S. P. Mukherjee, B. Lazzaretto, K. Hultenby, L. Newman, A. F. Rodrigues, N. Lozano, K. Kostarelos, P. Malmberg and B. Fadeel, Chem, 2018, 4, 334–358 CAS.
- S. P. Mukherjee, A. R. Gliga, B. Lazzaretto, B. Brandner, M. Fielden, C. Vogt, L. Newman, A. F. Rodrigues, W. Shao, P. M. Fournier, M. S. Toprak, A. Star, K. Kostarelos, K. Bhattacharya and B. Fadeel, Nanoscale, 2018, 10, 1180–1188 RSC.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan, H. Fang and R. Zhou, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef CAS PubMed.
- R. Li, N. D. Mansukhani, L. M. Guiney, Z. Ji, Y. Zhao, C. H. Chang, C. T. French, J. F. Miller, M. C. Hersam, A. E. Nel and T. Xia, ACS Nano, 2016, 10, 10966–10980 CrossRef CAS PubMed.
- Y. Zhang, W. Liu, Y. Li, Y. W. Yang, A. Dong and Y. Li, iScience, 2019, 19, 662–675 CrossRef CAS PubMed.
- X. Yang, J. Li, T. Liang, C. Ma, Y. Zhang, H. Chen, N. Hanagata, H. Su and M. Xu, Nanoscale, 2014, 6, 10126–10133 RSC.
- R. Wu, X. Ou, R. Tian, J. Zhang, H. Jin, M. Dong, J. Li and L. Liu, Nanoscale, 2018, 10, 20162–20170 RSC.
- K. Rasool, M. Helal, A. Ali, C. E. Ren, Y. Gogotsi and K. A. Mahmoud, ACS Nano, 2016, 10, 3674–3684 CrossRef CAS PubMed.
- M. Liang and X. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 CrossRef CAS PubMed.
- X. Zhang, S. Zhang, Z. Yang, Z. Wang, X. Tian and R. Zhou, Nanoscale, 2021, 13, 12613–12622 RSC.
- S. Chen, N. Shi, M. Huang, X. Tan, X. Yan, A. Wang, Y. Huang, R. Ji, D. Zhou, Y. G. Zhu, A. A. Keller, J. L. Gardea-Torresdey, J. C. White and L. Zhao, ACS Nano, 2021, 15, 16344–16356 CrossRef CAS PubMed.
- V. J. González, A. M. Rodríguez, I. Payo and E. Vázquez, Nanoscale Horiz., 2020, 5, 331–335 RSC.
- M. Himly, M. Geppert, S. Hofer, N. Hofstätter, J. Horejs-Höck and A. Duschl, Small, 2020, 16, e1907483 CrossRef PubMed.
- A. Khandagale, B. Lazzaretto, G. Carlsson, M. Sundin, S. Shafeeq, U. Römling and B. Fadeel, J. Leukocyte Biol., 2018, 104, 1199–1213 CrossRef CAS PubMed.
- C. Farrera, K. Bhattacharya, B. Lazzaretto, F. T. Andon, K. Hultenby, G. P. Kotchey, A. Star and B. Fadeel, Nanoscale, 2014, 6, 6974–6983 RSC.
- T. A. Fuchs, U. Abed, C. Goosmann, R. Hurwitz, I. Schulze, V. Wahn, Y. Weinrauch, V. Brinkmann and A. Zychlinsky, J. Cell Biol., 2007, 176, 231–241 CrossRef CAS PubMed.
- S. Patel, S. Kumar, A. Jyoti, B. S. Srinag, R. S. Keshari, R. Saluja, A. Verma, K. Mitra, M. K. Barthwal, H. Krishnamurthy, V. K. Bajpai and M. Dikshit, Nitric Oxide, 2010, 22, 226–234 CrossRef CAS PubMed.
- G. Peng, M. F. Montenegro, C. N. M. Ntola, S. Vranic, K. Kostarelos, C. Vogt, M. S. Toprak, T. Duan, K. Leifer, L. Bräutigam, J. O. Lundberg and B. Fadeel, Nanoscale, 2020, 12, 16730–16737 RSC.
- A. Manda-Handzlik, W. Bystrzycka, A. Cieloch, E. Glodkowska-Mrowka, E. Jankowska-Steifer, E. Heropolitanska-Pliszka, A. Skrobot, A. Muchowicz, O. Ciepiela, M. Wachowska and U. Demkow, Cell. Mol. Life Sci., 2020, 77, 3059–3075 CrossRef CAS PubMed.
- J. U. Lee, K. Kim and H. Cheong, 2D Mater., 2015, 2, 044003 CrossRef.
- C. Lee, H. Yan, L. E. Brus, T. F. Heinz, J. Hone and S. Ryu, ACS Nano, 2010, 4, 2695–2700 CrossRef CAS PubMed.
- S. Carlsson, M. Govoni, N. P. Wiklund, E. Weitzberg and J. O. Lundberg, Antimicrob. Agents Chemother., 2003, 47, 3713–3718 CrossRef CAS PubMed.
- Z. Zhuge, L. L. Paulo, A. Jahandideh, M. C. R. Brandao, P. F. Athayde-Filho, J. O. Lundberg, V. A. Braga, M. Carlström and M. F. Montenegro, Redox Biol., 2017, 13, 163–169 CrossRef CAS PubMed.
- S. Wu, Z. Zeng, Q. He, Z. Wang, S. J. Wang, Y. Du, Z. Yin, X. Sun, W. Chen and H. Zhang, Small, 2012, 8, 2264–2270 CrossRef CAS PubMed.
- A. Ambrosi, Z. Sofer and M. Pumera, Small, 2015, 11, 605–612 CrossRef CAS PubMed.
- A. Jiang, Y. Zhang, D. Wu, S. Li, Z. Liu, Z. Yang and Z. Wei, Ecotoxicol. Environ. Saf., 2021, 210, 111886 CrossRef CAS PubMed.
- S. Roy, A. Mondal, V. Yadav, A. Sarkar, R. Banerjee, P. Sanpui and A. Jaiswal, ACS Appl. Bio Mater., 2019, 2, 2738–2755 CrossRef CAS PubMed.
- J. Xu, R. Cai, Y. Zhang and X. Mu, Colloids Surf., B, 2021, 200, 111575 CrossRef CAS PubMed.
- A. Robert and B. Meunier, ACS Nano, 2022, 16, 6956–6959 CrossRef CAS PubMed.
- D. N. Douda, M. A. Khan, H. Grasemann and N. Palaniyar, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2817–2822 CrossRef CAS PubMed.
- K. Bhattacharya, C. Sacchetti, R. El-Sayed, A. Fornara, G. P. Kotchey, J. A. Gaugler, A. Star, M. Bottini and B. Fadeel, Nanoscale, 2014, 6, 14686–14690 RSC.
- G. Peng, T. Duan, M. Guo, Y. Xue, C. Chen, Y. Li, K. Leifer and B. Fadeel, Nanoscale, 2021, 13, 13072–13084 RSC.
- J. O. Lundberg, M. Carlström and E. Weitzberg, Cell Metab., 2018, 28, 9–22 CrossRef CAS PubMed.
- M. B. Lim, J. W. Kuiper, A. Katchky, H. Goldberg and M. Glogauer, J. Leukocyte Biol., 2011, 90, 771–776 CrossRef CAS PubMed.
- C. F. Urban, U. Reichard, V. Brinkmann and A. Zychlinsky, Cell. Microbiol., 2006, 8, 668–676 CrossRef CAS PubMed.
- B. Lazzaretto and B. Fadeel, J. Immunol., 2019, 203, 2276–2290 CrossRef CAS PubMed.
- M. Jiménez-Alcázar, C. Rangaswamy, R. Panda, J. Bitterling, Y. J. Simsek, A. T. Long, R. Bilyy, V. Krenn, C. Renné, T. Renné, S. Kluge, U. Panzer, R. Mizuta, H. G. Mannherz, D. Kitamura, M. Herrmann, M. Napirei and T. A. Fuchs, Science, 2017, 358, 1202–1206 CrossRef PubMed.
- M. J. Kaplan and M. Radic, J. Immunol., 2012, 189, 2689–2695 CrossRef CAS PubMed.
- V. Poli and I. Zanoni, Trends Microbiol., 2023, 31, 280–293 CrossRef CAS PubMed.
- D. Bender and G. Schwarz, FEBS Lett., 2018, 592, 2126–2139 CrossRef CAS PubMed.
- G. Mao, D. Xin, Q. Wang and D. Lai, Free Radicals Biol. Med., 2022, 182, 79–92 CrossRef CAS PubMed.
- M. Cao, R. Cai, L. Zhao, M. Guo, L. Wang, Y. Wang, L. Zhang, X. Wang, H. Yao, C. Xie, Y. Cong, Y. Guan, X. Tao, Y. Wang, S. Xu, Y. Liu, Y. Zhao and C. Chen, Nat. Nanotechnol., 2021, 16, 708–716 CrossRef CAS PubMed.
- G. Schwarz, R. R. Mendel and M. W. Ribbe, Nature, 2009, 460, 839–847 CrossRef CAS PubMed.
- L. B. Maia and J. J. Moura, Chem. Rev., 2014, 114, 5273–5357 CrossRef CAS PubMed.
- Y. Li, A. Yamaguchi, M. Yamamoto, K. Takai and R. Nakamura, J. Phys. Chem. C, 2017, 121, 2154–2164 CrossRef CAS.
- Z. Wang, A. von dem Bussche, Y. Qiu, T. M. Valentin, K. Gion, A. B. Kane and R. H. Hurt, Environ. Sci. Technol., 2016, 50, 7208–7217 CrossRef CAS PubMed.
- Y. Sang, W. Li, H. Liu, L. Zhang, H. Wang, Z. Liu, J. Ren and X. Qu, Adv. Funct. Mater., 2019, 29, 1900518 CrossRef.
- Z. Wang, Q. Tu, S. Zheng, J. J. Urban, S. Li and B. Mi, Nano Lett., 2017, 17, 7289–7298 CrossRef CAS PubMed.
- X. Cong, X. L. Liu, M. L. Lin and P. H. Tan, npj 2D Mater. Appl., 2020, 4, 13 CrossRef CAS.
- C. Farrera and B. Fadeel, J. Immunol., 2013, 191, 2647–2656 CrossRef CAS PubMed.
- K. Bhattacharya, C. Sacchetti, P. M. Costa, J. Sommertune, B. D. Brandner, A. Magrini, N. Rosato, N. Bottini, M. Bottini and B. Fadeel, Adv. Healthcare Mater., 2018, 7, e1700916 CrossRef PubMed.
- L. C. Potter, P. D. Millington, G. H. Thomas, R. A. Rothery, G. Giordano and J. A. Cole, FEMS Microbiol. Lett., 2000, 185, 51–57 CrossRef CAS PubMed.
- N. Hogg, Free Radicals Biol. Med., 2010, 49, 122–129 CrossRef CAS PubMed.
- J. Joseph, B. Kalyanaraman and J. S. Hyde, Biochem. Biophys. Res. Commun., 1993, 192, 926–934 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr03120a |
| This journal is © The Royal Society of Chemistry 2023 |