Open Access Article
Open Access ArticleUltrasensitive two-dimensional material-based MCF-7 cancer cell sensor driven by perturbation processes†
Sophia S. Y.
Chan
 a,
Denise
Lee
a,
Denise
Lee
 a,
Maria Prisca
Meivita
a,
Maria Prisca
Meivita
 a,
Lunna
Li
a,
Lunna
Li
 a,
Yaw Sing
Tan
a,
Yaw Sing
Tan
 b,
Natasa
Bajalovic
b,
Natasa
Bajalovic
 *a and
Desmond K.
Loke
*a and
Desmond K.
Loke
 *ac
*ac
aDepartment of Science, Mathematics and Technology, Singapore University of Technology and Design, Singapore 487372, Singapore. E-mail: natasa_bajalovic@sutd.edu.sg; desmond_loke@sutd.edu.sg
bBioinformatics Institute, Agency for Science, Technology and Research (A*STAR), Singapore 138671, Singapore
cOffice of Innovation, Changi General Hospital, Singapore, 529889, Singapore
First published on 25th October 2021
Abstract
Changes in lipid composition and structure during cell development can be markers for cell apoptosis or various diseases such as cancer. Although traditional fluorescence techniques utilising molecular probes have been studied, these methods are limited in studying these micro-changes as they require complex probe preparation and cannot be reused, making cell monitoring and detection challenging. Here, we developed a direct current (DC) resistance sensor based on two-dimensional (2D) molybdenum disulfide (MoS2) nanosheets to enable cancer cell-specific detection dependent on micro-changes in the cancer cell membrane. Atomistic molecular dynamics (MD) simulations were used to study the interaction between 2D MoS2 and cancer lipid bilayer systems, and revealed that previously unconsidered perturbations in the lipid bilayer can cause an increase in resistance. Under an applied DC sweep, we observed an increase in resistance when cancer cells were incubated with the nanosheets. Furthermore, a correlation was observed between the resistance and breast cancer epithelial cell (MCF-7) population, illustrating a cell population-dependent sensitivity of our method. Our method has a detection limit of ∼3 × 103 cells, below a baseline of ∼1 × 104 cells for the current state-of-the-art electrical-based biosensors using an adherent monolayer with homogenous cells. This combination of a unique 2D material and electrical resistance framework represents a promising approach for the early detection of cancerous cells and to reduce the risk of post-surgery cancer recurrence.
Introduction
Biomarkers are quintessential in healthcare to support the management of infectious diseases (recent examples include the use of antibodies and nucleic acids as biomarkers for Covid-19 tests),1 and to determine the severity and outcome of life-threatening diseases such as cancer. As one of the leading causes of death worldwide, cancer accounts for more than 9 million deaths per year with cases on the rise.2 Moreover, cancer is currently the leading cause of death in Singapore, accounting for approximately 28% of mortality in 2019.3 In particular, there is a need to discover a biomarker that can be used to detect diseases where early diagnosis is crucial or currently difficult. Changes in cell membrane components (e.g. lipids and proteins) are emerging as an ideal candidate for applications in diagnosis and treatment; for example, changes in lipid composition during cancer cell development can lead to carcinogenesis and metastasis.4,5 Detecting these cancer cell membrane-specific changes with high sensitivity is of vital significance in cancer treatments: for early cancer diagnosis to improve patient outcome, and to reduce the risk of cancer recurrence after cancer treatment.6,7Each cell type possesses a unique cell membrane composition, comprising a variety of phospholipids that form a bilayer, with embedded proteins.5,8,9 Phospholipids generally have a polar head group and a long, hydrophobic tail. Polar hydrophilic heads point outwards and are in contact with the aqueous fluids inside (cytoplasm) and outside (extracellular medium) the cell. On the other hand, the non-polar hydrophobic tails point inwards, and are not in contact with the aqueous phase inside and outside the cells. Typical phospholipid components found in the cell membrane are phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylethanolamine (PE), sphingolipid (SL) and cholesterol. The cell membrane is crucial for cell signalling and regulating cell processes (endocytosis, enzymatic activity, transport etc.), as well as its structural function.5,10 Normal cells have an asymmetric lipid bilayer, where the composition of the inner and outer layer, or leaflet are not the same.5,9 A loss in asymmetry can be associated with apoptosis or diseases such as cancer.5,11,12 Non-asymmetric cancer cell membranes tend to have different cell membrane potentials due to an increase in charged lipids (e.g. PS and PE) found in the outer leaflet.11,13,14 Changes in lipid asymmetry and composition contribute to the overall cancer cell membrane potential,15,16 and can be used to understand cellular mechanisms,17,18 or as biomarkers for cancer progression and detection,10,19,20 and targeted cancer-cell treatments.11,14
There are three different types of cell membrane potential: surface, transmembrane and dipole potential. The surface potential (Ψs) describes the accumulation of charges at the interface between the cell membrane and the extracellular media, while the transmembrane potential (ΔΨ) describes the difference in potential across the membrane. Cell membranes also have a dipole potential (Ψd), which defines the orientation of lipid dipolar residues (hydrophobic lipid tails) and extracellular water molecules.18,21,22 Surrounding water molecules, phospholipid heads and hydrophobic tails can contribute to changes in Ψd for cancer cell membranes due to the different lipid compositions and packing density.13,23–25 Changes in Ψd were also observed after lipid bilayer perturbation.26 Although lipid bilayer composition and disruption have been characterised via traditional fluorescent methods, they can be problematic because they tend to be indirect, or require additional steps for substrate preparation and usually cannot be reused.
Electrical-based techniques are advantageous and promising candidates for understanding perturbations in the cell membrane by investigating the changes in native bioelectrical signals.27–29 However, using traditional direct current (DC) or alternating current (AC) electrical stimulation to detect cancer cell types remains challenging due to poor sensitivity (a high limit of detection of several ten thousand cells). Advanced materials with unique electrical properties can improve the performance of the electrical-based devices.30,31 Recently, two dimensional (2D) materials have been used for cancer cell detection, including, graphene-based materials32,33 and transition metal dichalcogenides (TMDs).34,35 For example, devices based on functionalised graphene oxide have demonstrated high sensitivity for detecting circulating tumor cells (CTCs).36 In addition, a 2D graphite-like carbon nitride nanosheet-based electrode was used to detect cervical cancer cells (HeLa) in suspension with a low limit of detection. However, some of these nanomaterials suffer from long-term toxicity.37 Identifying an appropriate 2D material with a strong affinity for specific biomarkers, native biocompatibility and unique electrical properties is critical for developing a sensitive electrical-based cell detector.
Molybdenum disulfide (MoS2) is a TMD with unique physical, chemical, electrical and optical properties on a 2D scale.38,39 As the MoS2 thickness is reduced, the material transits from an indirect band gap in the bulk to a semiconductor with a direct band gap. MoS2 nanosheets are biocompatible,40,41 have a similar conductivity to cancer cells20 and are currently harnessed in biological applications such as cancer therapeutics (e.g. drug delivery and photothermal therapy),42–45 tissue engineering46,47 and biosensors20,48,49 and other applications. Although the electrical properties of MoS2 nanomaterials have been investigated, studies concerning the interaction between cancer cell membranes and MoS2, and its application in DC resistance sensing are limited.
Here, we explored the interactions between a cancer cell membrane and MoS2 nanosheet using atomistic molecular dynamics (MD) simulations. Our calculations demonstrated a previously unconsidered decrease in the Ψd after MoS2 interaction. We hypothesise that the decrease in Ψd is due to the lipid bilayer disruption and the final configuration of MoS2 within the lipid bilayer. Motivated by this theoretical finding, we developed an electrical resistance-based system using MoS2 and demonstrated a marked increase in resistance when epithelial breast cancer cells (MCF-7) were incubated with MoS2 nanosheets. Our experiments agreed well with our hypothesis: the addition of MoS2 nanosheets increased the resistance of the medium cell population. Furthermore, a correlation was observed between resistance and MCF-7 cell population, illustrating a cell population-dependent sensitivity of our method. We also demonstrate a limit of detection of ∼3 × 103 cells, lower than the baseline of ∼1 × 104 cells for current state-of-the-art electrical-based methods using adherent monolayer of homogenous cells, which allows for the detection of cells at low levels for clinically relevant cancer diagnosis and treatment. In addition, our system preparation uses time-saving few steps and all the components can be reused compared to traditional time-consuming multi-step detection systems, allowing for a rapid diagnosis while saving cost. This proposed combination of a unique 2D material and electrical resistance framework is a promising method for the development of lab-on-chip platforms for clinical purposes.
Experimental
Simulation parameterisation
MoS2 sheet was prepared as previously described.50 A 4.1 nm × 4.2 nm single-layer MoS2 sheet with two zig-zag edges, one terminating with S atoms and the other with Mo atoms, and two armchair edges was generated using Materials Studio. Bonded force field parameters were provided by Varshney et al.,51 while non-bonded force field parameters were provided by Luan and Zhou52 (see ESI Table S2†). Potential energy contributions from bond angles larger than 82° in the MoS2 sheet were not considered.A model lipid bilayer system representing the cell membrane of a cancer cell was generated using CHARMM-GUI.53,54 The lipid bilayer system was composed of neutral dipalmitoylphosphatidylcholine (DOPC) and negatively charged dipalmitoylphosphatidylserine (DOPS) lipids (composition found in ESI Table S3†). There were 242 lipids in total, 121 in each leaflet, and the system was solvated in a solution with 40 TIP3P water molecules per lipid and 0.15 M NaCl.55 The system was equilibrated for 50 ns before monolayer MoS2 was added in subsequent simulation runs.
Simulation details
Atomistic MD simulations were performed with GROMACS 5.0.4 using the Slipids force field as described before.50,55–57 Initially, the MoS2 nanosheet was positioned parallel with and ∼2.5 nm above the lipid bilayer. The simulation box, with dimensions of 8.72 nm × 8.86 nm × 10.50 nm, was filled with 1341 TIP3P water molecules,58 0.15 NaCl, and 52 sodium counterions. The system was energy minimised using the steepest descent algorithm for up to 2000 steps, and then equilibrated in the isochoric–isothermal (NVT) ensemble for t = 100 ps. The temperature of the system was maintained at T = 300 K using the velocity-rescaling thermostat59 with a coupling constant of 0.5 ps. Another equilibration step was performed in the isothermal–isobaric (NPT) ensemble for t = 100 ps, followed by the production run for 700 ns. The pressure was kept constant by the Parrinello–Rahman barostat at P = 1 bar,60 with a semi-isotropic pressure coupling scheme while the temperature was kept constant using the Nosé–Hoover thermostat.61,62 Two independent MD simulation runs with different initial atomic velocities were performed. Long-range electrostatic interactions were treated with the particle-mesh Ewald scheme,63,64 and van der Waals interactions were calculated with a cut-off distance of 1 nm. All lipid and MoS2 bonds were constrained using the LINCS algorithm. A time step of t = 2 fs was used with a leapfrog integrator and coordinates of all atoms were saved every 20 ps.Data analysis
Interaction energies between the MoS2 and the lipid bilayer were extracted from the simulation using the Gromacs tool gmx energy. They were obtained as Lennard-Jones potentials, which are van der Waals interactions.Using the Gromacs tool gmx mindist, atom count was calculated for the number of lipid atoms close to the MoS2 nanosheet. The total atom count describes the total number of lipid atoms that are less than 6 Å away from the MoS2 nanosheet at any time during the simulation run.
Transmembrane dipole potential (Ψd) was calculated using the Gromacs tool gmx potential. The potential was calculated from the charge density, which was obtained from one hundred slice profiles along the x–y plane. The thickness of each slice was ∼0.92 Å. The dipole potential was obtained by double integration of charge density using the following equation:13,25
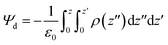 | (1) |
The angle of the MoS2 nanosheet relative to the lipid bilayer's x–y plane was calculated for every frame of the trajectory using the Gromacs tool gmx bundle. The angle was between the nanosheet and z-axis was measured for every frame (20 ps) during the trajectory, and it was then translated to the lipid bilayer axis by 90°.
TMD preparation
Few layer MoS2 sheets in deionised water (2D Semiconductors) were sonicated (Elmasonic P) for ∼10 min at room temperature (RT) before material characterisation and incubation. MoS2 was drop casted on silicon (Si) substrate prior to the Raman spectroscopy and scanning electron microscopy (SEM). The chemical composition of the 2D MoS2 sheets were analysed by energy-dispersive X-ray spectroscopy (EDS) point analysis: the EDS analysis indicated the stoichiometric ratio of Mo![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S to be about 1
S to be about 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.
2.
Cell culture
Breast cancer cell line MCF-7 were cultured in Dulbecco's Modified Eagle Media (DMEM) (Nacalai Tesque) containing 7% fetal bovine serum (FBS) and 5 mM of L-glutamine (Gibco), maintained in a 5% CO2 environment at 37 °C. Cells were observed using an inverted light microscope with a 10× objective lens (Olympus DP22).Electrical characterisation
MCF-7 cells (3 × 103 cells) were seeded in our setup 24 h prior to material addition; ITO electrodes deposited on a glass substrate (LaTech) with a cloning cylinder (Sigma) adhered with silicone adhesive (Fig. 2a). After incubating the material with the cells for an additional 24 h, current–voltage (I–V) sweeps (V = 0 V to −5 V, dual sweep) were performed using the semiconductor characterisation system (Keithly 4200-SCS). The resistance (R) was calculated for all the voltage points using the following equation: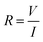 | (2) |
Cell viability assay
To investigate MoS2 cytotoxicity, MCF-7 cells were plated in 96-well plates and cultured ∼24 h prior to treatment. Sonicated MoS2 nanosheets were added at a concentration ranging from 0–100 μM at 25 μM increments for t = 24 h and 48 h. Cell viability and cytotoxic effects of MoS2 nanosheets were determined using a WST-1 assay. After adding 10% WST-1 (Sigma) in media, the cells were incubated for ∼4 h at 37 °C. Cell viability was estimated by measuring absorbance at λ = 450 nm. Cell viability was also tested ∼24 h after applying the DC sweep.Results & discussion
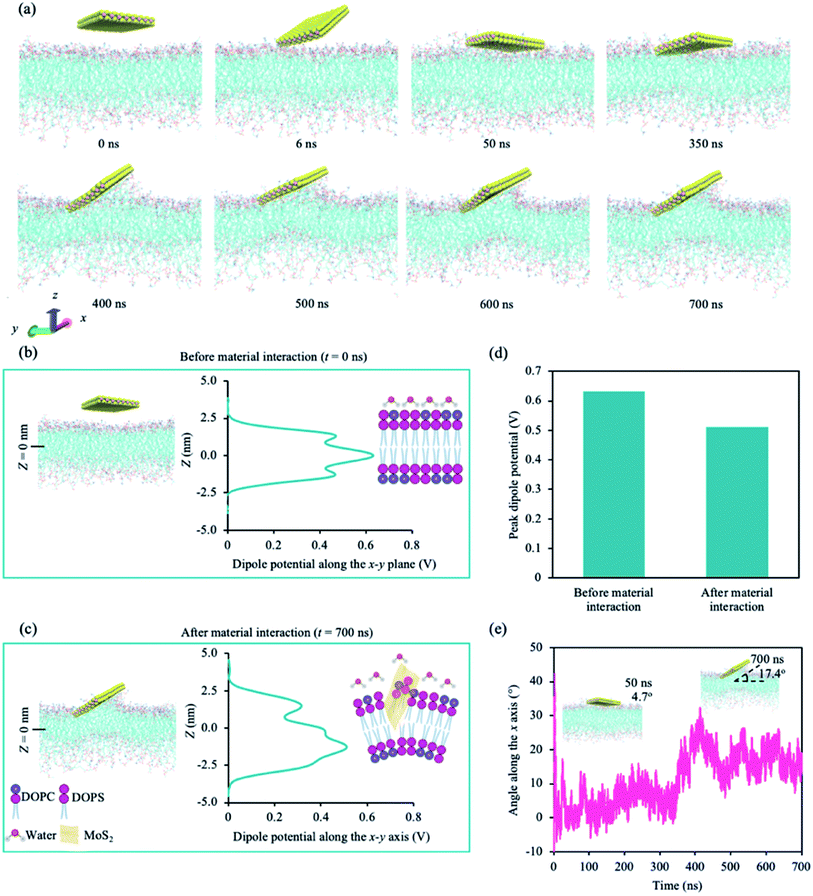
We performed atomistic molecular dynamic (MD) simulations to understand the interaction between the cancer lipid bilayer system and a MoS2 nanosheet. Two simulation runs were performed for 700 ns each, and we observed similar interactions between MoS2 and the lipid bilayer for both runs. Fig. 1 shows representative data obtained from one of the two simulation runs. Our model lipid bilayer for the cell membrane of a cancer cell is a simplified version of that modelled by Klähn and Zacharias,65 wherein the outer lipid layer contains an increased proportion of phosphatidylserine (PS) compared to that of a healthy cell (lipid composition can be found in ESI Table S2†).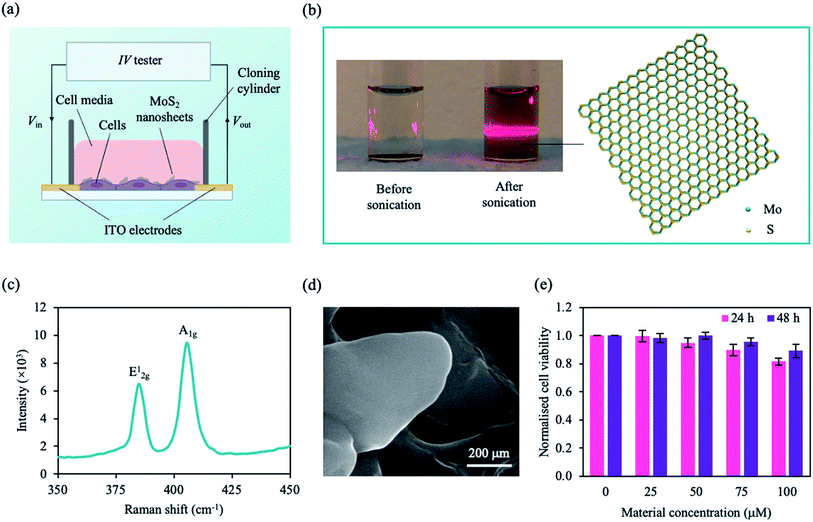 | ||
| Fig. 2 Configuration of our system and MoS2 characterisation. (a) A schematic of our setup. Cells were plated onto the ITO electrodes on a glass substrate and incubated with MoS2 nanosheets. Using an IV tester, a DC sweep was applied and the cell signal was measured. (b) Photograph of MoS2 solution before and after sonication (left panel). Upon laser irradiation, the Faraday–Tyndall effect was only observed for the sonicated MoS2 solution as indicated by the strong red line through the solution. Schematic illustration of an MoS2 nanosheet with a hexagonal S–Mo–S structure (right panel). Colour coding of species: S, yellow; Mo, cyan. (c) Raman spectra the MoS2 nanosheet showing two vibrational modes. (d) Scanning electron microscope (SEM) image of MoS2 nanosheets deposited on silicon (Si) substrate. (e) Normalised cell viability plotted as a function of MoS2 concentration and incubation time (24 h, pink; 48 h, purple). Cell viability was estimated using a WST-1 assay with measured absorbance at λ = 450 nm. MCF-7 cells were grown in 96 well plates for ∼24 h prior to treatment. Data represent mean ± SEM (n = 6 from 3 independent experiments). See ESI Table S1† for significance. | ||
Fig. 1a shows the snapshots of the simulation with time. The first point of contact is between a corner of the MoS2 nanosheet and the upper leaflet of the lipid bilayer (∼6 ns). The nanosheet then tilts towards the lipid bilayer, aligning itself parallel to the lipid bilayer at around 50 ns. At ∼400 ns, the nanosheet is tilted, and phospholipids are extracted onto the sulphur (S) side facing the lipid bilayer (phospholipid extraction). Moreover, the lipid bilayer appears to curve below the MoS2 nanosheet, suggesting the disruption of the lipid bilayer. The same corner is in contact with the lipid bilayer during the entire simulation. We also calculated the changes in atom count (the number of lipid atoms less than 6 Å away from the MoS2 nanosheet) and van der Waals (vdW) interaction energy (Fig. S1a†). Two significant changes were observed for atom count and vdW energy: one around 50 ns and the other at around 400 ns, which correspond to the nanosheet laying parallel to the upper leaflet of the lipid bilayer and the tilting of the nanosheet on the lipid bilayer respectively (Fig. S1a†). The center of mass (CoM) distance was also calculated between the nanosheet and the upper leaflet of the lipid bilayer (Fig. S1b†). After the initial contact, CoM was generally consistent during the simulation.
We calculated the dipole potential (Ψd) of the cancer lipid bilayer system before and after MoS2 interaction (Fig. 1b and c). Without MoS2, the system has a Ψd profile that is typical of a pure lipid bilayer system, consistent with literature (Fig. 1b).13,25 Using a reference value of 0 V for the aqueous phase, the profile showed a sharp increase in Ψd across the lipid head region with a peak value of ∼0.47 V. These peaks were located ∼1.28 nm (Z = ±1.28 nm) above and below the bilayer center (Z = 0 nm), corresponding to the top part of the lipid tails near the lipid head groups. The local minima, typically associated with the unsaturated double bonds along the lipid tails, were observed ∼0.8 nm (Z = ±0.8 nm) above and below the bilayer center. A global maximum of ∼0.63 V was reached at Z = 0 nm, which can be attributed to the presence of partial changes and corresponding dipoles at the terminal methyl groups on the lipid tails. After MoS2 interaction, the Ψd profile of the lipid bilayer system becomes asymmetric (Fig. 1c): the Ψd profile broadens, the global maximum decreases (∼0.51 V) and the peak shifts downwards from z = 0 nm to −1.5 nm (Fig. 1b–d). This could be due to lipid bilayer perturbation, where the curvature of the lipid bilayer induced by the MoS2 nanosheet can result in the majority of the terminal methyl groups shifting away from the bilayer center. There is a new peak located at ∼1.4 nm above from the bilayer center (Z = +1.4 nm), which could be associated with phospholipid extraction as some of the lipids shift away from the bilayer center towards the material. Moreover, the angle of MoS2 relative to the lipid bilayer (x–y) plane was calculated for the duration of the simulation (Fig. 1e). As observed in Fig. 1a, the MoS2 nanosheet aligns almost parallel (∼4.7°) to the lipid bilayer before tilting towards the end of the simulation (∼17.4°) (Fig. 1e). The vdW interactions between the MoS2 nanosheet perturbed the lipid bilayer and resulted in bilayer curvature and phospholipid extraction (Fig. 1a–c), causing the changes in the Ψd profile, and could be considered as added resistance to the system (interlayer resistance).66–68 This is because current prefers to flow laterally through the material (along the nanosheet).66 To enhance the current flow, the ideal orientation of MoS2 is parallel to the cell membrane. As a result, it is likely that the current is impeded by the perturbation of the lipid bilayer and tilted orientation of the MoS2 nanosheet relative to the lipid bilayer.
We adopted an MoS2-based DC resistance system to detect a medium population of MCF-7 cells based on micro-changes in the cell membrane. Our system comprises of two 650 nm-thick indium tin oxide (ITO) left and right electrodes deposited on a glass substrate, and a cloning cylinder was adhered to the electrodes using a silicone adhesive (Fig. 2a). The measured current is sensitive to the changes in Ψd due to the micro-changes in the cell membrane with the addition of the MoS2 nanosheets. As embedded MoS2 can perturb the lipid bilayer and affect Ψd, MCF-7 cells can be detected based on the changes in the resistance in the presence of the nanosheets.
Before interacting with the cells, 2D MoS2 nanosheets were obtained by liquid-phase sonication to break the vdW forces between the layers. Each MoS2 nanosheet consists of a few hexagonal S–Mo–S sheets (Fig. 2b) held together via vdW interactions. The structure and morphology of the sonicated samples were investigated. The nanosheets were well dispersed within the solution as observed by the typical red beam through the sonicated MoS2 solution under irradiated light (Faraday–Tyndall effect) (Fig. 2b). MoS2 flakes were drop casted on a silicon (Si) substrate prior to material characterisation. The Raman spectra (Fig. 2c) shows two characteristic Raman vibrational modes of the few-layered MoS2, labelled E12g at ∼384.80 cm−1 and A1g at ∼405.46 cm−1, similar to experimental data in literature.69 The degenerate modes of E12g symmetry correspond to the in-plane transversal and longitudinal vibrations of the sublayers in opposite directions, while the A1g symmetry is a result of the third mode opposite-in-phase out-of-plane vibrations of the sublayers.69Fig. 2d shows a scanning electron microscope (SEM) image of the 2D MoS2 flake, in which thin nanosheets were observed.
To investigate the interaction of the cell with MoS2, we characterised the cytotoxicity of MoS2 nanosheets for MCF-7 cells at different material concentrations and incubation times (Fig. 2e). Cell viability was determined using WST-1 cell proliferation assay (see Experimental section). After ∼24 h, cell viability decreased with increasing material concentration. However, after ∼48 h of incubation, cell viability was comparable to that of control (cells only). We concluded that, within our tested concentration range, the material is not cytotoxic to MCF-7 cells after ∼48 h. To maximise our current output while maintaining cell viability, we chose a nanosheet concentration of 75 μM. MCF-7 cells were not significantly affected by MoS2 at ∼75 μM after 48 h (Fig. 2e), consistent with observations in previous studies.41,70
To study the micro-changes in the cell membrane as a result of MoS2 interaction, we investigated the cell-dependent resistance of MCF-7 cells incubated with MoS2 nanosheets (MCF-7/MS) and control (cells only) using a DC current–voltage (I–V) sweep with a negative voltage bias (V = 0 V to −5 V). The average resistance for MCF-7/MS was calculated using eqn (2) (see Experimental section), and normalised with control. Fig. 3a depicts a material-dependent increase in resistance obtained for MCF-7 cells. MCF-7/MS demonstrated an increase in normalised resistance compared to control (∼9%, 3 × 103 cells), confirming the decrease in native Ψd as a result of adding MoS2 nanosheets. Based on the results of our MD simulations, we speculate that the perturbation of the lipid bilayer system (resulting from the embedded MoS2) and the final angle of the nanosheet contributed to the increase in resistance. Since current flows laterally (along the nanosheet) of the MoS2, the tilt of the nanosheet disrupts the current flow along the cell membrane. Moreover, the extracted phospholipids could have increased the resistance between the nanosheet and the lipid bilayer, acting as an insulator and impeding current flow. Additionally, Fig. 3a shows the resistance for MCF-7/MS at different cell populations normalised to control of each cell population. After plating cells at different cell populations within our setup (n = 3 × 103, 4 × 103 and 5 × 103 cells), we observed a significant increase in normalised resistance for MCF-7/MS plated at 4 × 103 and 5 × 103 cells. While it is expected that resistance should increase with cell population,71–73 we observed no significant difference in normalised resistance across controls plated at different cell populations (3 × 103, 4 × 103 and 5 × 103 cells) (ESI Fig. S2†). On the other hand, the system showed a ∼22% and ∼28% increase in the normalised resistance when cell population was increased from 3 × 103 to 4 × 103 cells and 5 × 103 cells for MCF-7/MS, respectively. We determined the sensitivity of detection for our system by calculating the percentage change in normalised resistance between MCF-7/MS and control (ΔR%)
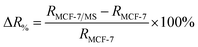 | (3) |
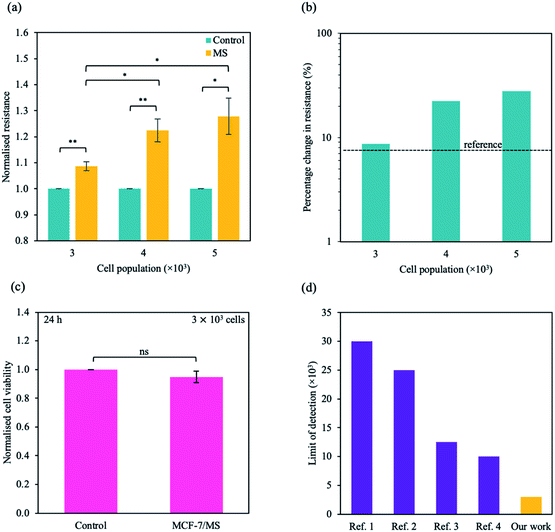 | ||
| Fig. 3 Sensing micro-changes in the MCF-7 cell membrane using an MoS2-based DC resistance sensor. (a) Normalised resistance for control (cells only) (green) and MCF-7/MS (yellow) plotted for various cell populations (n = 3 × 103, 4 × 103 and 5 × 103 cells). The resistance was calculated from the DC sweep with a voltage bias of V = 0 to −5 V, and resistance values were normalised to its own control. (b) Semi-logarithmic plot of percentage change in normalised resistance for different cell populations. The black dotted line represents the reference resistance change (background signal + three times the standard deviation of the background signal (σ)). (c) Normalised cell viability plotted as a function of the presence of material. Cell viability was determined ∼24 h after DC sweep using WST-1 assay measured at λ = 450 nm. (d) A plot of limit of detection for systems detecting adherent cells developed by other groups (purple) and this work presented here (yellow). Full references are available in ESI Table S4.† Data in (a) and (c) represent mean ± SEM (n = 6 from 3 independent experiments). Significance was calculated using a Student's t-test: non-significant (ns), p < 0.05 (*), p < 0.01 (**). For (a), unlabelled significance is denoted as ns. | ||
Due to the clinical importance of cell detection, numerous works have focused on developing an fluorescent-based method to detect various cell types (e.g. cancer cells, stem cells).76–78 While there have been some promising research in detecting low numbers of cells in suspension, traditional fluorescent methods can require additional functionalisation, complex device preparation and additional time for result output.79 Our work describes a few-step method to detect an adherent, single cell type without complex preparation methods, with a facile set up that can be combined with an electrical circuit. Here, we describe a method to detect an adherent monolayer of cancer cells using MoS2-based DC resistance sensor. Using our method, we demonstrate a limit of detection of ∼3 × 103 cells, below the baseline of ∼1 × 104 cells for current state-of-the-art electrical-based biosensors with an adherent monolayer of homogenous cells.80–83
Alternative methods to traditional fluorescence detection such as electrochemical and electrical-based devices are promising for detecting various, specific cell types. Conventional electrochemical methods have been used as immunosensors with low limits of detection.30,84,85 However, traditional electrochemical methods are primarily indirect and based on sensing biomolecules eliminated from cells. In addition, these methods can rely on substrates with a high degree of functionalisation for cell or biomolecule-specific detection. Preparation of these substrates can be complex and time consuming, and might not be reusable. On the other hand, our system is capable of detecting the cells directly without needing additional functionalisation steps, reducing the complexity of our method. The components of our proposed setup can also be easily removed, sterilised and reused, greatly reducing the cost of our system for easy adoption.
Although impedance sampling can be performed on 3D scaffolds (tissues) or cells, sample preparation and measurement are more complex for tissue samples. Excised tissues are typically non-homogenous, containing various cell types with varying shapes, sizes and functions.86,87 Moreover, the organisation and architecture of cells can vary between tissue samples, resulting in different measured outputs between each tissue slice or sample. Electrical characterisation is further complicated by the extracellular matrix that varies within tissue types.87,88 As a result, performing electrical-based characterisation on a 2D cell monolayer can avoid the above complications. Furthermore, characterisations conducted in cells can be useful to understand the response of a single cell type, cell dynamics or cell-specific treatments.
On a cellular level, electrical characterisation can be performed for single-cells, cells in suspension and adhered cells in a monolayer. However, traditional methods of manipulating cells for single-cell measurements can be complex and cell-to-cell variations may result in inconsistency. Although results obtained from cells in suspension can be informative, it is difficult to translate the results to the tissue level as they cannot mimic the natural environment of tissues (e.g. cell–cell interactions, extracellular matrix etc.). Furthermore, cells in isolation do not respond as well to electric fields due to the lack of biochemical cues.71,87,89 While a 2D cell monolayer lacks in vivo imitation, our method does not require complex sample preparation and can produce results within a shorter period of time.86 Cell monolayers represent great potential for understanding the cell behaviour within a homogenous population, which can be valuable for clinical applications (e.g. cell-specific detection, drug testing). We present a few-step solution based on the combination of 2D MoS2 and DC resistance to detect adherent cancer cells with a low limit of detection.
Conclusion
It has been challenging to detect an adherent monolayer of cells using electrical-based biosensors. Here, we demonstrate a sensitive detection method for a small adherent population of MCF-7 cells utilising an MoS2-based DC resistance sensor. Atomistic MD simulations illustrate a decrease in Ψd due to MoS2-facilitated lipid bilayer perturbation. Using our method, we were able to detect these micro-changes in the cell membrane with high sensitivity and low detection limit. Our proposed methodology represents a building block towards the development of clinically relevant lab-on-chip platforms. Nevertheless, further investigations for high throughput are needed for successful clinical and practical applications of the MoS2-based electrical resistance sensor.Conflicts of interest
The authors declare no competing interests.Acknowledgements
We thank C. Cheung, K. G. Lim, L. T. Ng for important discussions. The authors acknowledge support from the Singapore University of Technology and Design (SUTDT12017003), Changi General Hospital (Singapore) (CGH-SUTD-HTIF2019-001), Ministry of Education (Singapore) (MOE2017-T2-2-064), SUTD-Zhejiang-University (SUTD-ZJU (VP) 201903) grant programs and Agency of Science, Technology and Research (Singapore) (A20G9b0135) grant programs. D. K. L. acknowledges support from the Massachusetts Institute of Technology–SUTD International Design Centre and National Supercomputing Centre, Singapore (15001618). S. S. Y. Chan acknowledges support from the SUTD President Graduate Scholarship.References
- T. Ji, Z. Liu, G. Q. Wang, X. Guo, S. Akbar khan, C. Lai, H. Chen, S. Huang, S. Xia, B. Chen, H. Jia, Y. Chen and Q. Zhou, Biosens. Bioelectron., 2020, 166, 112455 CrossRef CAS PubMed.
- National Cancer Institute, Cancer Statistics – National Cancer Institute, https://www.cancer.gov/about-cancer/understanding/statistics, accessed 3 May 2021.
- A. Lim, 10 Important Cancer Statistics in Singapore (Updated 2021), https://smartwealth.sg/cancer-statistics-singapore/, accessed 28 June 2021 Search PubMed.
- L. M. Butler, Y. Perone, J. Dehairs, L. E. Lupien, V. de Laat, A. Talebi, M. Loda, W. B. Kinlaw and J. V. Swinnen, Adv. Drug Delivery Rev., 2020, 159, 245–293 CrossRef CAS PubMed.
- S. Zalba and T. L. M. ten Hagen, Cancer Treat. Rev., 2017, 52, 48–57 CrossRef CAS PubMed.
- T. Li, Q. Fan, T. Liu, X. Zhu, J. Zhao and G. Li, Biosens. Bioelectron., 2010, 25, 2686–2689 CrossRef CAS PubMed.
- H. A. Parsons, J. Rhoades, S. C. Reed, G. Gydush, P. Ram, P. Exman, K. Xiong, C. C. Lo, T. Li, M. Fleharty, G. J. Kirkner, D. Rotem, O. Cohen, F. Yu, M. Fitarelli-Kiehl, K. W. Leong, M. E. Hughes, S. M. Rosenberg, L. C. Collins, K. D. Miller, B. Blumenstiel, L. Trippa, C. Cibulskis, D. S. Neuberg, M. DeFelice, S. S. Freeman, N. J. Lennon, N. Wagle, G. Ha, D. G. Stover, A. D. Choudhury, G. Getz, E. P. Winer, M. Meyerson, N. U. Lin, I. Krop, J. C. Love, G. M. Makrigiorgos, A. H. Partridge, E. L. Mayer, T. R. Golub and V. A. Adalsteinsson, Clin. Cancer Res., 2020, 26, 2556–2564 CrossRef CAS PubMed.
- S. J. Singer and G. L. Nicolson, Science, 1972, 175, 720–731 CrossRef CAS PubMed.
- F. M. Goñi, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 1467–1476 CrossRef PubMed.
- M. R. Wenk, Nat. Rev. Drug Discovery, 2005, 4, 594–610 CrossRef CAS PubMed.
- L. T.-H. Tan, K.-G. Chan, P. Pusparajah, W.-L. Lee, L.-H. Chuah, T. M. Khan, L.-H. Lee and B.-H. Goh, Front. Pharmacol., 2017, 8, 1–6 CAS.
- G. L. Nicolson, Biochim. Biophys. Acta, Biomembr., 2014, 1838, 1451–1466 CrossRef CAS PubMed.
- G. Shahane, W. Ding, M. Palaiokostas and M. Orsi, J. Mol. Model., 2019, 25, 1–13 CrossRef PubMed.
- T. Rivel, C. Ramseyer and S. Yesylevskyy, Sci. Rep., 2019, 9, 1–14 CAS.
- A. A. Marino, I. G. Iliev, M. A. Schwalke, E. Gonzalez, K. C. Marler and C. A. Flanagan, Tumor Biol., 1994, 15, 82–89 CrossRef CAS PubMed.
- R. Latorre and J. E. Hall, Nature, 1976, 264, 361–363 CrossRef CAS PubMed.
- M. Yang and W. J. Brackenbury, Front. Physiol., 2013, 4, 185 CAS.
- T. Starke-Peterkovic, N. Turner, P. L. Else and R. J. Clarke, Am. J. Physiol.: Regul., Integr. Comp. Physiol., 2005, 288, R663–R670 CrossRef CAS PubMed.
- S. Eissa, R. Chinnappan and M. Zourob, Electroanalysis, 2017, 29, 1994–2000 CrossRef CAS.
- Y. Chen, J. Peng, Y. Lai, B. Wu, L. Sun and J. Weng, Biosens. Bioelectron., 2019, 142, 111520 CrossRef CAS PubMed.
- H. Brockman, Chem. Phys. Lipids, 1994, 73, 57–79 CrossRef CAS PubMed.
- L. Wang, Annu. Rev. Biochem., 2012, 81, 615–635 CrossRef CAS PubMed.
- R. F. Flewelling and W. L. Hubbel, Biophys. J., 1996, 49, 541–552 CrossRef.
- R. A. Pearlstein, C. J. Dickson and V. Hornak, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 177–194 CrossRef CAS PubMed.
- W. Ding, M. Palaiokostas, W. Wang and M. Orsi, J. Phys. Chem. B, 2015, 119, 15263–15274 CrossRef CAS PubMed.
- M. Jan Akhunzada, F. D'Autilia, B. Chandramouli, N. Bhattacharjee, A. Catte, R. Di Rienzo, F. Cardarelli and G. Brancato, Sci. Rep., 2019, 9, 1–12 CAS.
- Y. Furukawa, A. Shimada, K. Kato, H. Iwata and K. Torimitsu, Biochim. Biophys. Acta, Gen. Subj., 2013, 1830, 4329–4333 CrossRef CAS PubMed.
- S. K. Rastogi, J. Bliley, D. J. Shiwarski, G. Raghavan, A. W. Feinberg and T. Cohen-Karni, Cell. Mol. Bioeng., 2018, 11, 407–418 CrossRef CAS PubMed.
- M. Mayer, O. Arrizabalaga, F. Lieb, M. Ciba, S. Ritter and C. Thielemann, Biosens. Bioelectron., 2018, 100, 462–468 CrossRef CAS PubMed.
- D. K. Loke, G. J. Clausen, J. F. Ohmura, T. C. Chong and A. M. Belcher, ACS Appl. Nano Mater., 2018, 1, 6556–6562 CrossRef CAS.
- L. Li, A. M. Belcher and D. K. Loke, Nanoscale, 2020, 12, 24214–24227 RSC.
- L. Feng, Y. Chen, J. Ren and X. Qu, Biomaterials, 2011, 32, 2930–2937 CrossRef CAS PubMed.
- Y. Song, Y. Chen, L. Feng, J. Ren and X. Qu, Chem. Commun., 2011, 47, 4436–4438 ( Chem , 2020 , 311 , 127863 ) RSC.
- Y. Shu, L. Zhang, H. Cai, Y. Yang, J. Zeng, D. Ma and Q. Gao, Sens. Actuators, B, 2020, 311, 127863 CrossRef CAS.
- X. Qian, S. Shen, T. Liu, L. Cheng and Z. Liu, Nanoscale, 2015, 7, 6380–6387 RSC.
- H. J. Yoon, T. H. Kim, Z. Zhang, E. Azizi, T. M. Pham, C. Paoletti, J. Lin, N. Ramnath, M. S. Wicha, D. F. Hayes, D. M. Simeone and S. Nagrath, Nat. Nanotechnol., 2013, 8, 735–741 CrossRef CAS PubMed.
- L. Cheng, X. Wang, F. Gong, T. Liu and Z. Liu, Adv. Mater., 2020, 32, 1902333 CrossRef CAS PubMed.
- M. Xu, T. Liang, M. Shi and H. Chen, Chem. Rev., 2013, 113, 3766–3798 CrossRef CAS PubMed.
- M. Chhowalla, Z. Liu and H. Zhang, Chem. Soc. Rev., 2015, 44, 2584–2586 RSC.
- J. H. Appel, D. O. Li, J. D. Podlevsky, A. Debnath, A. A. Green, Q. H. Wang and J. Chae, ACS Biomater. Sci. Eng., 2016, 2, 361–367 CrossRef CAS PubMed.
- S. S. Chan, Y. S. Tan, K. X. Wu, C. Cheung and D. K. Loke, ACS Appl. Bio Mater., 2018, 1, 210–215 CrossRef CAS.
- X. Zhu, X. Ji, N. Kong, Y. Chen, M. Mahmoundi, X. Xu, L. Ding, W. Tao, T. Cai, Y. Li, T. Gan, A. Barret, Z. Bharwani, H. Chen and O. C. Farokhzad, ACS Nano, 2018, 12, 2922–2938 CrossRef CAS PubMed.
- S. Cai, J. Yan, H. Xiong, Q. Wu, H. Xing, Y. Liu, S. Liu and Z. Liu, Int. J. Pharm., 2020, 590, 119948 CrossRef CAS PubMed.
- T. Liu, C. Wang, W. Cui, H. Gong, C. Liang, X. Shi, Z. Li, B. Sun and Z. Liu, Nanoscale, 2014, 6, 11219–11225 RSC.
- B. Pang, H. Yang, L. Wang, J. Chen, L. Jin and B. Shen, Colloids Surf., A, 2021, 608, 125506 CrossRef CAS.
- S. Wang, J. Qiu, W. Guo, X. Yu, J. Nie, J. Zhang, X. Zhang, Z. Liu, X. Mou, L. Li and H. Liu, Adv. Biosyst., 2017, 1, 1–9 Search PubMed.
- H. Nazari, A. Heirani-Tabasi, M. S. Alavijeh, Z. S. Jeshvaghani, E. Esmaeili, S. Hosseinzadeh, F. Mohabatpour, B. Taheri, S. H. A. Tafti and M. Soleimani, ChemistrySelect, 2019, 4, 11557–11563 CrossRef CAS.
- J. Lee, P. Dak, Y. Lee, H. Park, W. Choi, M. A. Alam and S. Kim, Sci. Rep., 2014, 4, 1–7 Search PubMed.
- A. Kálosi, M. Labudová, A. Annušová, M. Benkovičová, M. Bodík, J. Kollár, M. Kotlár, P. Kasak, M. Jergel, S. Pastoreková, P. Siffalovic and E. Majkova, Biomater. Sci., 2020, 8, 1973–1980 RSC.
- S. S. Y. Chan, Y. S. Tan, K.-X. Wu, C. Cheung and D. K. Loke, ACS Appl. Bio Mater., 2018, 1, 210–215 CrossRef CAS.
- V. Varshney, S. S. Patnaik, C. Muratore, A. K. Roy, A. A. Voevodin and B. L. Farmer, Comput. Mater. Sci., 2010, 48, 101–108 CrossRef CAS.
- B. Luan and R. Zhou, Appl. Phys. Lett., 2016, 108, 1–5 CrossRef.
- S. Jo, T. Kim, V. G. Iyer and W. Im, J. Comput. Chem., 2008, 29, 1859–1865 CrossRef CAS PubMed.
- S. Jo, J. B. Lim, J. B. Klauda and W. Im, Biophys. J., 2009, 97, 50–58 CrossRef CAS PubMed.
- J. P. M. Jämbeck and A. P. Lyubartsev, J. Chem. Theory Comput., 2012, 8, 2938–2948 CrossRef PubMed.
- J. P. M. Jämbeck and A. P. Lyubartsev, J. Phys. Chem. B, 2012, 116, 3164–3179 CrossRef PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindah, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- W. G. Hoover, Phys. Rev. A: At., Mol., Opt. Phys., 1985, 31, 1695–1697 CrossRef PubMed.
- S. Nosé, J. Chem. Phys., 1984, 81, 511–519 CrossRef.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- M. Klähn and M. Zacharias, Phys. Chem. Chem. Phys., 2013, 15, 14427–14441 RSC.
- S. Das and J. Appenzeller, Nano Lett., 2013, 13, 3396–3402 CrossRef CAS PubMed.
- J. Na, M. Shin, M. K. Joo, J. Huh, Y. Jeong Kim, H. Jong Choi, J. Hyung Shim and G. T. Kim, Appl. Phys. Lett., 2014, 104, 233502 CrossRef.
- S. Das and J. Appenzeller, Phys. Status Solidi RRL, 2013, 7, 268–273 CrossRef CAS.
- L. Liang and V. Meunier, Nanoscale, 2014, 6, 5394–5401 RSC.
- P. Shah, T. N. Narayanan, C. Z. Li and S. Alwarappan, Nanotechnology, 2015, 26, 1–7 CrossRef CAS PubMed.
- P. Shah, X. Zhu, X. Zhang, J. He and C. Z. Li, ACS Appl. Mater. Interfaces, 2016, 8, 5804–5812 CrossRef CAS PubMed.
- Y. An, T. Jin, F. Zhang and P. He, J. Electroanal. Chem., 2019, 834, 180–186 CrossRef CAS.
- L. Wang, J. Zhu, C. Deng, W. L. Xing and J. Cheng, Lab Chip, 2008, 8, 872–878 RSC.
- H. Wang, H. Li, Y. Huang, M. Xiong, F. Wang and C. Li, Biosens. Bioelectron., 2019, 142, 111531 CrossRef CAS PubMed.
- J. Liu, Y. Qin, D. Li, T. Wang, Y. Liu, J. Wang and E. Wang, Biosens. Bioelectron., 2013, 41, 436–441 CrossRef CAS PubMed.
- I. R. Suhito, N. Angeline, S. S. Choo, H. Y. Woo, T. Paik, T. Lee and T. H. Kim, Sensors, 2018, 18, 1–17 CrossRef PubMed.
- J. Zhang, J. Shi, W. Liu, K. Zhang, H. Zhao, H. Zhang and Z. Zhang, Sens. Actuators, B, 2018, 276, 552–559 CrossRef CAS.
- D. Yang, M. Liu, J. Xu, C. Yang, X. Wang, Y. Lou, N. He and Z. Wang, Talanta, 2018, 185, 113–117 CrossRef CAS PubMed.
- Z. Liao, L. Han, Q. Li, L. Li, Y. Liu, Y. Song, W. Tan and E. Song, Adv. Funct. Mater., 2021, 31, 1–11 Search PubMed.
- C. H. Yea, H. C. Jeong, S. H. Moon, M. O. Lee, K. J. Kim, J. W. Choi and H. J. Cha, Biomaterials, 2016, 75, 250–259 CrossRef CAS PubMed.
- H. C. Jeong, S. S. Choo, K. T. Kim, K. S. Hong, S. H. Moon, H. J. Cha and T. H. Kim, Sens. Actuators, B, 2017, 242, 224–230 CrossRef CAS.
- I. R. Suhito, E. S. Kang, D. S. Kim, S. Baek, S. J. Park, S. H. Moon, Z. Luo, D. Lee, J. Min and T. H. Kim, Colloids Surf., B, 2019, 180, 384–392 CrossRef CAS PubMed.
- N. Angeline, S. S. Choo, C. H. Kim, S. H. Bhang and T. H. Kim, BioChip J., 2021, 15, 52–60 CrossRef CAS.
- Q. Guo, X. Li, C. Shen, S. Zhang, H. Qi, T. Li and M. Yang, Microchim. Acta, 2015, 182, 1483–1489 CrossRef CAS.
- A. A. M. Abdurhman, Y. Zhang, G. Zhang and S. Wang, Anal. Bioanal. Chem., 2015, 407, 8129–8136 CrossRef CAS PubMed.
- M. Kapałczyńska, T. Kolenda, W. Przybyła, M. Zajączkowska, A. Teresiak, V. Filas, M. Ibbs, R. Bliźniak, Ł. Łuczewski and K. Lamperska, Arch. Med. Sci., 2018, 14, 910–919 Search PubMed.
- M. J. Peters, J. G. Stinstra and I. Leveles, in Modeling and Imaging of Bioelectrical Activity, 2004, pp. 281–319 Search PubMed.
- H. P. Schwan, Adv. Biol. Med. Phys., 1957, 5, 147–209 CAS.
- K. Zhu, N. R. Hum, B. Reid, Q. Sun, G. G. Loots and M. Zhao, Sci. Rep., 2020, 10, 1–11 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Simulation results and other experimental data. See DOI: 10.1039/d1na00614b |
| This journal is © The Royal Society of Chemistry 2021 |