Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Structural motifs and intramolecular interactions in non-canonical G-quadruplexes
Jagannath
Jana
,
Swantje
Mohr
,
Yoanes Maria
Vianney
 and
Klaus
Weisz
and
Klaus
Weisz
 *
*
Institute of Biochemistry, Universität Greifswald, Felix-Hausdorff-Str. 4, D-17487 Greifswald, Germany. E-mail: weisz@uni-greifswald.de; Fax: +49 3834 420-4427; Tel: +49 3834 420-4426
First published on 22nd January 2021
Abstract
Guanine(G)-rich DNA or RNA sequences can assemble or intramolecularly fold into G-quadruplexes formed through the stacking of planar G·G·G·G tetrads in the presence of monovalent cations. These secondary nucleic acid structures have convincingly been shown to also exist within a cellular environment exerting important regulatory functions in physiological processes. For identifying nucleic acid segments prone to quadruplex formation, a putative quadruplex sequence motif encompassing closely spaced tracts of three or more guanosines is frequently employed for bioinformatic search algorithms. Depending on the number and type of intervening residues as well as on solution conditions, such sequences may fold into various canonical G4 topologies with continuous G-columns. On the other hand, a growing number of sequences capable of quadruplex formation feature G-deficient guanine tracts, escaping the conservative consensus motif. By folding into non-canonical quadruplex structures, they adopt unique topologies depending on their specific sequence context. These include G-columns with only two guanines, bulges, snapback loops, D- and V-shaped loops as well as interlocked structures. This review focuses on G-quadruplex species carrying such distinct structural motifs. It evaluates characteristic features of their non-conventional scaffold and highlights principles of stabilizing interactions that also allow for their folding into stable G-quadruplex structures.
Introduction
Single-stranded guanine-rich DNA or RNA sequences can fold into intramolecular or intermolecular four-stranded structures called G-quadruplexes (G4s). G4-prone motifs are found in high numbers not only in bacterial and viral, but also in human genomes. Thus, G-rich oligonucleotides derived from genomic sequences like those from oncogene promoters and telomeres have been demonstrated to fold into G-quadruplexes. Through their visualization, compelling evidence for the existence of these non-canonical secondary nucleic acid structures has also been found in cellular environments.1,2 Our current understanding of the biological roles of quadruplexes suggests that G4s are involved in gene regulation and telomere maintenance, making genomic quadruplexes promising therapeutic targets.3 In this regard, much effort has been devoted during the last decades to searching for G4-stabilizing ligands for pharmaceutical intervention, e.g., for modulating gene expression or telomerase inhibition in cancer cells.4 In addition to serving as potential drug targets, synthetic quadruplexes such as the thrombin binding aptamer (TBA) or anti-HIV-1 integrase aptamer constitute an emerging class of therapeutics, binding to various molecules including many pathologically relevant proteins with very high affinity and selectivity.5,6 Finally, the increasing use of quadruplexes in supramolecular chemistry as well as in biosensors and nanotechnology as a result of their ability to self-organize into complex two-dimensional networks and long nanowires attests to their enormous potential in medicinal and technological applications.7–9A typical monomolecular G-quadruplex is formed by sequences harboring four G-tracts of three or more consecutive guanosine residues separated by short intervening sequences. Correspondingly, conservative search algorithms are based on a consensus sequence motif d(G3+N1−7G3+N1−7G3+N1–7G3+) for predicting putative G4 structures in genomic DNA.10,11 However, a growing number of non-consensus sequences has been reported to actually fold into stable G4 species. The availability of their high-resolution structures has shown a variety of unique conformational features distinct from the ‘classical’ G4 architecture. Clearly, a better understanding of principles governing quadruplex folding of such non-standard G-rich sequences will support new algorithms for predicting putative regions within the genome amenable to G4 formation,12,13 but may also expand the G4 structural landscape for more effective drug targeting or the engineering of novel G4-based scaffolds.
This review is primarily focusing on the increasing number of G4 structures that do not comply with a consensus sequence motif but rather rely on short G2-tracts and/or isolated G nucleotides for their architecture. Various strategies to compensate for G-deficiencies within their G-core or for reduced stacking interactions between tetrads are surveyed to give more insight into relevant contributions to G4 stability. Given the large number of deposited G4 structures with unusual sequence motifs, emphasis is placed on the folding behavior of unmodified sequences, with less attention given to quadruplexes featuring several closely spaced tracts of four or more consecutive guanosines and non-canonical tetrads, i.e., those composed of additional residues other than Gs.
A short survey on canonical G-quadruplex structures
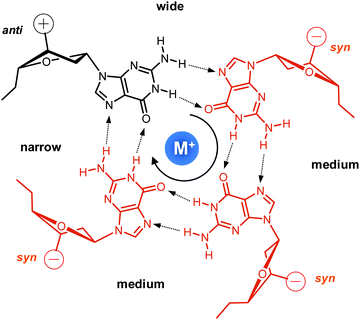
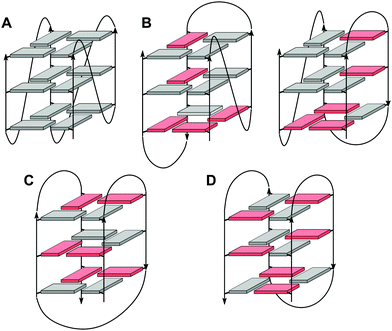
Upon folding of a sequence composed of four closely spaced GGG triplets, guanine bases from the G-tracts will associate to form planar G-quartets (G-tetrads) through a cyclic hydrogen bond pattern involving both their Hoogsteen and Watson–Crick faces (Fig. 1). In most cases, stacking of three G-tetrads gives a three-layered G-core that is additionally stabilized through monovalent cations with a strength of stabilization in the order K+ > Na+ ≥ NH4+ > Li+.14 These are coordinated within the central channel of the G-core that is lined by the G-carbonyl oxygens to create a strong negative potential.In case of an intramolecular quadruplex, intervening sequences form loop regions connecting the four G-columns (Fig. 2). A propeller or double-chain-reversal loop links two adjacent G-tracts with parallel orientation whereas lateral (edge-wise) and diagonal loops connect two adjacent or distal anti-parallel G-tracts, respectively. Depending on its topology, a conventional monomolecular quadruplex may be grouped into three major families: a parallel G4 with all four G-tracts being parallel and only containing propeller loops; an anti-parallel G4 with two parallel and two anti-parallel G-runs; and a (3+1) hybrid with three parallel and one anti-parallel G-columns. Because an intramolecular quadruplex is defined by a combination of three different types of loops progressing in either a clockwise or counter-clockwise direction, a large number of topologies is conceivable. For a more systematic nomenclature, a descriptor based on the type of consecutive loops and their progression in relation to a frame of reference has been suggested.15 In such a system, the parallel topology with three sequential propeller loops progressing in an anti-clockwise direction can be designated as (–p–p–p) (Fig. 2A). Clearly, several of the theoretical loop combinations are forbidden due to geometrical restrictions. In fact, only 14 of these were predicted to be mechanically feasible but four of those have still not been experimentally verified to-date.16,17
Among conformational properties of individual G residues within the quadruplex core, glycosidic torsion angles, i.e. syn and anti conformers, play a critical role for any quadruplex species due to their importance in G-tetrad formation and their close link with relative strand polarities of the four G-columns. In a parallel quadruplex, all residues within a tetrad must adopt the same glycosidic torsion angle for forming a planar G-quartet arrangement held together by the eight Hoogsteen hydrogen bond interactions. Typically, such G4s are composed of an all-anti G-core, although exceptions forming a single all-syn quartet exist for modified but also unmodified quadruplexes.18,19 On the other hand, residues in anti-parallel G-tracts require different glycosidic conformations when participating in the same G-tetrad. This relationship between relative strand polarities and glycosidic torsion angles has frequently been used as a powerful tool to guide folding of a G-quadruplex through the site-specific incorporation of G analogs favoring either syn or anti glycosidic torsion angles to enforce a particular topology.
Whereas the pattern of glycosidic angles for residues within a G-tetrad is determined by the orientation of the four G-columns, the sequential glycosidic conformation of consecutive G residues within an individual G-run may vary. It should be noted, however, that changing the glycosidic torsion angle within a column will, as a consequence, also change the tetrad polarity, i.e., the clockwise or anti-clockwise direction of Hoogsteen hydrogen bonds within a tetrad plane when going from hydrogen bond donor to hydrogen bond acceptor. Thus, anti–anti and syn–syn steps will result in homopolar tetrad stacking whereas syn–anti and anti–syn steps will lead to heteropolar stacking. Computational studies have predicted more favorable interactions for syn–anti and anti–anti steps with energetic penalties for anti–syn and syn–syn steps, consistent with conformational properties of most reported G-quadruplex structures.20,21
In addition to their relationship with relative strand orientation and G-tetrad polarity, glycosidic torsion angles will also affect the width of the four grooves featured by the four-stranded quadruplex. Whereas all grooves in parallel quadruplexes are of medium width, base-paired Gs of different glycosidic conformation as observed in anti-parallel and (3+1) hybrid structures will also form narrow and wide grooves in case of syn → anti and anti → syn arrangements within a tetrad, respectively.
From a perspective of intervening sequences, it is their folding into a specific type of loop that defines the topology of the quadruplex architecture. General guidelines have emerged, correlating the length and position of linker sequences to the stability and to favored G4 structures.22–24 Due to geometric restraints, formation and stability of particular loops are strongly correlated with the length of the intervening linker sequences. Generally, propeller loops are most stable when composed of only 1–2 residues although even 0 nt propeller loops have been reported in rare cases.25,26 Lateral loops often include 2–4 residues depending on bridging a narrow or a wide groove, and diagonal loops require ≥3 residues for linking diagonally positioned G nucleotides. However, even for a conventional sequence, additional complexity may arise due to loop and overhang residues being engaged in subtle tertiary interactions to likewise affect the favored topology. Finally, it is not only the inherent sequence but also the outer conditions like the nature of cations, the ionic strength, and molecular crowding that may significantly impact the topology of a folded quadruplex. Whereas sodium ions have been shown to promote an anti-parallel topology, potassium ions rather tend to destabilize anti-parallel quadruplexes.14,27 Folding of the same sequence into either a monomolecular or bimolecular quadruplex may depend on low or high potassium ion concentrations in the buffer solution and is yet another example for an often rather unpredictable folding pathway even when looking at regular G4-forming sequences.28,29
Taken together, intense research during the past years has provided a wealth of information regarding the energetics and structural interdependencies in ‘conventional’ G-quadruplexes. Our present knowledge of folding principles enables us to make a guess as for the most stable quadruplex fold of a given G4 consensus sequence and to tackle the rational design of G4 architectures.30,31 However, we are still far from reliably predicting topologies based on primary structure due to more subtle additional interactions involving flanking and intervening residues and also to the impact of specific solution conditions.
Quadruplexes with long loops and quadruplex–duplex hybrids
Longer unstructured loops in G-quadruplex structures tend to be increasingly disfavored because of entropic effects.32–34 In fact, only few quadruplexes with long loops of >7 residues, violating the conservative consensus sequence for putative G-quadruplex forming motifs, have been reported to-date. Thus, a G-quadruplex formed by the conserved 26 nt G-rich fragment of the human CEB25 minisatellite forms a parallel-stranded G-quadruplex with a 9 nt central double-chain-reversal loop (Fig. 3A).35 Within this quadruplex, an A·T Watson–Crick and a potential G·A non-canonical base pair between loop and 5′-overhang residues fix the 3′-terminal loop domain above the 5′-outer G-tetrad. Another example involves two parallel-stranded G4 conformers from a KRAS promoter sequence, that were found to coexist in equilibrium and feature long third propeller loops composed of eleven and twelve nucleotides, respectively. In this case, high-resolution NMR structures determined from single mutants revealed π–π interactions between some bases of the propeller loop as contributors to the overall stability of the structure.36 Also, sequences encompassing five to seven human telomeric (GGGTTA) repeats were shown by NMR to form (3+1) hybrid structures with an up to 21 nt long propeller loop when inner GGG triplets were blocked from participation in G-tetrads through single G → I or G → T substitutions.37 Noticeably, such long loops may constitute new recognition motifs, allowing their targeting by a loop-complementary oligonucleotide to form a double-helical loop region.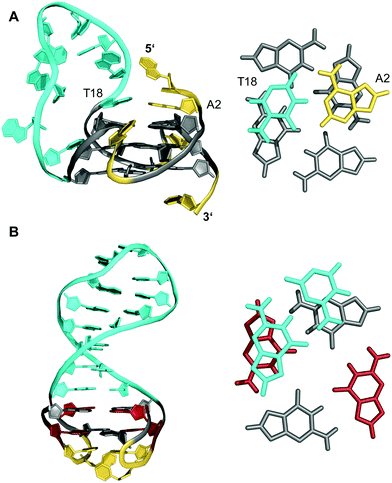 | ||
| Fig. 3 (A) Solution structure of a human CEB25 minisatellite sequence with a 9 nt propeller loop formed in K+ solution (20 mM KPi + 70 mM KCl, pH 7.0; Tm = 76.5 °C; PDB 2LPW);35 an A·T Watson–Crick base pair between a loop and 5′-overhang residue anchors the 3′-terminus of the loop on top of the 5′-outer G-tetrad (right). (B) Solution structure of a quadruplex–duplex hybrid with a two-layered anti-parallel G-quadruplex and a coaxially stacked duplex hairpin bridging the G4 wide groove formed in K+ solution (20 mM KPi + 20 mM KCl, pH 7.0; PDB 2M8Z);38 the duplex GC base pair stacks onto the G-tetrad at the quadruplex–duplex interface (right); anti- and syn-guanosines of the G-core as well as loop and flanking residues are colored grey, red, and yellow, respectively; the 9 nt propeller loop in (A) and the stem-loop duplex in (B) are colored cyan. | ||
Contrary to what is expected assuming most stable 1 nt propeller loops,39 longer loops of ≥5 residues are rather frequently found to be of a propeller type. Apparently, such loops often allow for stabilizing tertiary interactions with other loop and flanking residues. Following the concept of loop interactions to stabilize longer loop domains, appropriate linker sequences may intrinsically form Watson–Crick paired stem-loop duplexes as part of stable quadruplex–duplex hybrid structures (QDHs). Notably, in contrast to a regular single-stranded linker, quadruplex stabilities of engineered QDHs generally increase with the length of the double-helical hairpin domain.40 When forming a lateral-type loop connection, the duplex is favored to bridge a wide groove of the quadruplex G-core to better accommodate distances between the sugar-phosphate backbones of coaxially oriented duplex and quadruplex domains (Fig. 3B).31,38 Here, the first base pair at the junction also affects stability due to additional stacking interactions with the quadruplex outer tetrad.40 On the other hand, a connecting hairpin element may likewise replace a regular propeller loop, but by connecting G residues at opposite faces of the G-core the first base pair bridging the junction will be invariably disrupted in such an orthogonal arrangement.
Likewise, a duplex-forming diagonal loop with intrinsic Watson–Crick base pairing can be found for a G-rich sequence located in a promoter region of the HIV-1 long terminal repeat (LTR).41 In the major G-quadruplex conformation LTR-III, the 12 nt loop contains a stabilizing duplex hairpin element with three base pairs. However, the longer distance across the distal edges of the quadruplex again prevents residues at the quadruplex–duplex interface to be engaged in a stable base pair.
Quadruplexes with a two-tetrad G-core
The stability of G-quadruplexes is mostly derived from the stacking of its planar tetrads with stacking energies estimated to be ∼80 kJ mol−1 per tetrad.42,43 Therefore, the stability increases with an increase of stacked tetrads and only a limited number of monomeric two-layered quadruplex architectures has been reported to date. Among these, the thrombin-binding DNA aptamer (TBA) with its four tracts of only two contiguous Gs is one of the most prominent representatives.44–46 Each of its four G-tracts has a favorable 5′-syn–anti-3′ arrangement resulting in opposite hydrogen bond directionalities of its two stacked G(syn)–G(anti)–G(syn)–G(anti) tetrads. The G-runs are connected by two T–T lateral loops on one side and a central T–G–T lateral loop on the other side of the anti-parallel chair-type quadruplex (Fig. 4). Additional stabilization comes from the stacking of a TT base pair from the first and third loop on one of the G-quartets. Other stabilizing contributions may also involve some stacking interactions by bases of the central 3 nt lateral loop on the other face of the G-quadruplex core.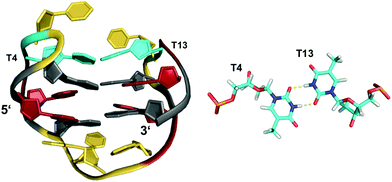 | ||
| Fig. 4 Chair-type anti-parallel G-quadruplex of the TBA aptamer d(GGTTGGTGTGGTTGG) in K+ solution (110 mM KCl, pH 6.1) and hydrogen-bonded base pair formed between two T residues from opposite T–T lateral loops (PDB 148D);46anti- and syn-Gs of the quadruplex core, loop residues, and the T·T base pair are colored grey, red, yellow, and cyan, respectively. | ||
Various TBA modifications have been reported in the past, mostly aiming at an improvement of pharmacological properties. Notably, a TBA analog containing a 5′–5' site of polarity inversion in the first lateral loop resulted in a (3+1) hybrid structure by keeping a 5′-syn–anti-3′ torsion angle progression along all G-runs.47 As a consequence, it differs from the unmodified TBA in having one G(syn)–G(syn)–G(syn)–G(anti) and one G(anti)–G(anti)–G(anti)–G(syn) tetrad alignment with a parallel 5′–3′ strand orientation of the first, second, and fourth strand and a third strand proceeding in the opposite direction. However, stabilizing forces through loop residues are very similar to those found for unmodified TBA.
The TBA quadruplex illustrates a frequently observed principle of stabilization through capping structures formed by base pairing alignments of loop and overhang residues. Such interactions may even be favorable enough in two-layered quadruplexes to successfully compete with three-layered G4s in sequences comprising four GGG-tracts. Thus, the unexpected observation of a G-quadruplex with only two tetrad layers for a human telomeric sequence featuring four G3-runs emphasizes the potential role of tertiary interactions.48,49 Usually, the human telomeric sequence exhibits a (3+1) hybrid form with three stacked G-tetrads in K+ solution. However, the 5′-truncated variant d[(GGGTTA)3GGGT] was shown to favor a two-layered basket-type structure with all G-columns comprising a 5′-syn–anti-3′ glycosidic bond arrangement.48 The conformation is stabilized by A·G·A and G·G·G triples capping the top and bottom faces of the G-core, respectively (Fig. 5). Moreover, two hydrogen-bonded T residues on top of each triad may add further stacking interactions.
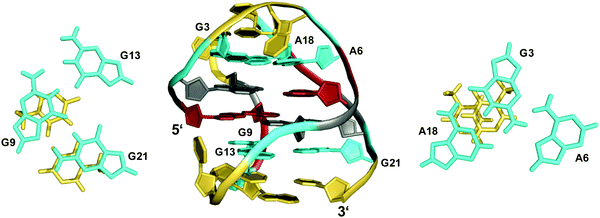 | ||
| Fig. 5 Structure of a human telomeric G-quadruplex (form 3) in K+ solution (20 mM KPi + 70 mM KCl, pH 7.0; Tm = 57.0 °C; PDB 2KF8);48 A18·G3·A6 and G21·G9·G13 base triads sandwiched between a G-tetrad and a potential T·T base pair cap the top and bottom of the two-layered G-core; anti- and syn-residues of the G-quadruplex core, loop and flanking residues, and bases involved in triads are colored grey, red, yellow, and cyan, respectively. | ||
Apparently, extensive base pairing and stacking of loop residues can outweigh stabilities of alternate three-layered G4 structures. It should be mentioned, however, that the telomeric two-G-tetrad conformation has been questioned to be a stable form for the extended human telomeric sequence because the addition of a 5′-flanking residue was shown to mostly abolish formation of a two-layered G4 structure.49 Instead, the latter was suggested to likely constitute an intermediate in the interconversion between different telomeric G-quadruplex topologies.
In close analogy to the human telomeric sequence mentioned above, a sequence with single mutation from the RANKL gene d(G3TAG3AGCG3AGAG3) adopts a two-layered basket-type topology, again stabilized by a G·G·G and an A·G·A triple on top of the 5′- and 3′-tetrad, respectively.50 Here, the critical role of capping base triads and loop residues was uncovered by a structural rearrangement to the anticipated three-layered (3+1) hybrid fold induced by an A5-to-T5 modification. The latter is associated with the destruction of the capping A5·G3·A17 triple, releasing G3 from the A5·G3 base pair. This enables G3 to participate in G-tetrad formation with a concomitant shortening of the 3 nt lateral loop to become a more favorable 2 nt propeller loop.
Other examples exist for the stabilization of a two-layered quadruplex core by additional tiers of planar base pairing arrangements from overhang and loop residues.51–54 Thus, a truncated form of the Bombyx mori telomeric single repeat sequence d(TAGG) was shown to fold into a four-stranded quadruplex with a two-fold symmetry axis consisting of two G(syn)–G(syn)–G(anti)–G(anti) tetrads of different tetrad polarity.51 The two-layered core is sandwiched between unusual T·A·A triads with one adenosine pairing with the A–T Watson–Crick pair through the minor groove. All three bases and the sugar ring of one adenosine of the triad partially stack over the underlying G residues of the quartet. Inspired by the latter architecture, a sequence d(GGGTTCAGG) was designed and demonstrated to fold into a two-fold symmetric bimolecular G4 structure with heteropolar stacking of two G(syn)–G(anti)–G(syn)–G(anti) tetrads capped by a C·G·A triad on each of the two quadruplex faces.52 Emphasizing the important role of additional layers made up by triads, the 12mer sequence d(A2G2T4A2G2) with a pair of AAGG repeats folds into a bimolecular structure with 2-fold symmetry and a core of two G(syn)–G(syn)–G(anti)–G(anti) tetrads capped on both sides by A·T·A triads.53 The latter, sandwiched between a G-tetrad and an additional outer non-Watson–Crick A–T base pair, contains one adenosine in syn conformation that pairs with the thymine through a reverse Hoogsteen alignment (Fig. 6). It should be mentioned that synergistic effects between the unusual base triads and the G4 core result in significant contributions of the stacked triads to the stability of two-layered quadruplexes but also to the promotion of base triad formation through the tetrad platform.
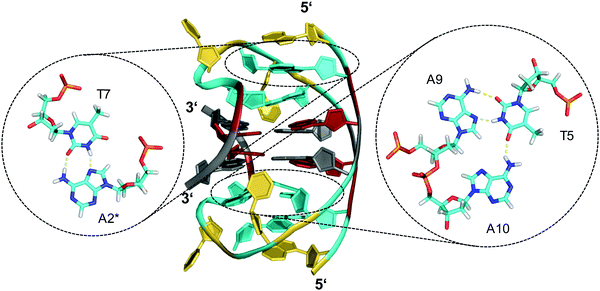 | ||
| Fig. 6 Diamond-shaped bimolecular G-quadruplex with a two-layered G-core formed in Na+ solution (5 mM NaPi + 150 mM NaCl, pH 6.9; PDB 1D6D);53 each of the tetrads forms a platform that is capped by a T(anti)·A(syn)·A(anti) triad (right) and a reversed Hoogsteen A·T base pair (left); an asterisk denotes a residue from the symmetry-related strand; anti- and syn-residues of the G-quadruplex core, loop and flanking residues, and bases involved in triads and base pairs are colored grey, red, yellow, and cyan, respectively. | ||
A 12 nt minimal sequence d[GT(GGT)3G] derived from the anti-proliferating 28 nt DNA aptamer AGRO100 forms a unique left-handed parallel G-quadruplex with two G-tetrad layers connected by short loops (Fig. 7A).55 Lacking additional capping structures, it dimerizes through 5′–5′ stacking for additional stabilization (Fig. 7B). Likewise, two monomers connected by a linker form a four-layered structure with two stacked left-handed subunits of parallel topology. Single-residue loops are clearly favored for the formation of the left-handed G4. In fact, thymine bases of the 1 nt loops collapse toward the terminal G-tetrad and allow for hydrogen bonds between their O4′ atoms and amino protons of adjacent tetrad guanines (Fig. 7C). Whereas the TBA sequence features four GG doublets, the 12 nt sequence of the left-handed G4 comprises two single Gs at each terminus. By their stacking upon each other they form an unusual split-guanine tract which is assumed to convey the left-handed twist with its fully circling backbone (Fig. 7C).
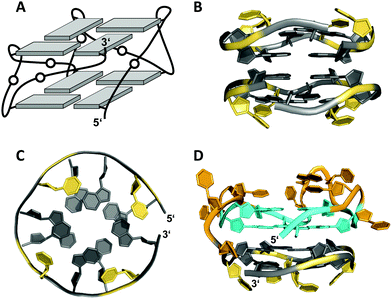 | ||
| Fig. 7 (A) Schematic representation of a minimal left-handed G4. (B) Crystal structure with two stacked left-handed G4 units (crystals grown from 12 mM spermine and 80 mM KCl, pH 7.0; PDB 6FQ2).55 (C) Top view with T loop residues oriented towards the outer tetrad of the left-handed domain; a broken G-column is formed by two split Gs at the 5′- and 3′-ends. (D) Hybrid structure with a right-handed TBA subunit connected to the left-handed motif formed in K+ solution (20 mM KPi + 70 mM KCl, pH 7.0; PDB 6JCE).56 G residues of the quadruplex core and loop residues are colored grey and yellow for the left-handed G4, and cyan and orange for the TBA subunit in (D). | ||
Apparently, parallel-stranded two-layered quadruplexes from sequences that encompass closely spaced G-doublets have a strong propensity for additional stacking interactions, either through dimerization or in case of longer sequences through a stacked arrangement of two G4 domains made up of their 5′- and 3′-segments. Several examples for the latter derive from the polymorphic AGRO100 aptamer that is composed of two domains with four G2-tracts each. A single G-to-T substitution in the 5′-stretch and addition of thymidine residues at the termini yielded a well-defined sequence that folds into a four-layered G-quadruplex comprising two propeller-type parallel-stranded subunits connected through a central linker.57 On the other hand, an alternate G-to-T substitution in the 3′-terminal G-doublet yielded a quadruplex topology termed Z-G4, featuring two stacked G4 domains both with left-handed helicity.58 Noticeably, the latter is enforced by the 3′-domain composed of the 12 nt minimal motif mentioned above.55
The TBA sequence can also be forced into a parallel topology with its three lateral loops switching into three propeller loops by its linkage to the minimal left-handed G4 sequence. Here, the two G4 units again stack on each other, yet with different helical orientation (Fig. 7D).56 Because lateral loops impede stacking, favorable stacking interactions between the two subunits, i.e., the right-handed TBA and the left-handed domain are efficient in driving such refolding into a parallel G4. Also, additional stacking of one base from each propeller loop on the 3′-outer TBA tetrad was observed and may contribute to the stabilization of this TBA topology.
Stabilization can also be provided by bases that are directly linked in-plane to the G-tetrad to form pentads, hexads or heptads. Thus, a dimeric hexad motif with two hexads stacking upon each other was reported for a d(GGAGGAG) sequence in a 150 mM Na+ solution.59 GGA triplet repeats are abundant in eukaryotic genomes and thought to also be associated with the occurrence of several diseases.60,61 In the two tandem GGA triplet repeat sequence, each bimolecular monomer forms a stack of a G·(A)·G·G·(A)·G hexad, a G-tetrad, and an A·A mismatched base pair (Fig. 8). The hexad forms by the in-plane attachment of two adenine bases over their Hoogsteen edge to the G-tetrad through hydrogen bonding with opposite guanine bases. Thus, two out of the four G-tetrad guanines are anchored through a total of six hydrogen bonds. Formation of such hexads is expected to be supported or even driven by extensive π–π stacking interactions between two stacked hexads at the dimer interface.
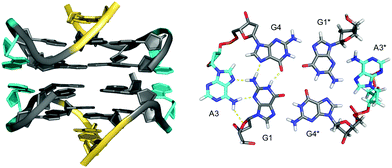 | ||
| Fig. 8 Dimeric G4 structure formed in Na+ solution (2 mM NaPi + 150 mM NaCl, pH 6.6) and composed of four symmetry-related strands with stacked hexads at the dimer interface (PDB 1EEG);59 guanosines of the all-anti quadruplex core, loop and flanking residues, and adenosines involved in hexads are colored grey, yellow and cyan, respectively. | ||
A corresponding architecture with an intramolecular stack composed of a G·(A)·G·(A)·G·(A)·G heptad and a G-tetrad, additionally stabilized through dimer formation with stacked heptads at the interface, was also found for a four tandem GGA triplet repeat d(GGA)4. Likewise, an intramolecularly folded d(GGA)8 extended sequence with two subunits composed of stacked tetrad and heptad arranged in a tail-to-tail orientation with inter-heptad stacking.62,63 Notably, although octad formation through the G-tetrad alignment of a fourth adenine base either from the 3′-terminus in d(GGA)4 or from the adenosine linking the two subunits in d(GGA)8 is conceivable, it has not been observed. Apparently, the adenosine requires a subsequent 3′-adjacent G residue as part of the G-tetrad to be anchored within the tetrad plane.
In conclusion, the structure of two-layered quadruplexes as presented above emphasize the need for other stabilizing interactions in addition to the stacking of two tetrads in a G-core composed of favorable syn–anti or anti–anti steps along the four GG-columns. Here, loop residues are of particular importance by forming base pairs and/or base triads as additional stacked layers sandwiching the G-quadruplex core. Also, dimerization or inter-subunit stacking is often observed in case of two-layered quadruplexes with a propeller-type parallel topology, enabling unrestricted stacking with interfacial 5′-outer tetrads generally found to be more favorable.64 Stacking interactions can be further optimized by expanding G-tetrads with intervening bases to form hexads or heptads, increasing the stacking interface within dimeric structures.
G-deficient G-quadruplexes and interrupted G-tracts
In the past, an increasing number of G4-forming sequences harboring a shortened G-tract and thus unable to fold into a canonical three-layered quadruplex with four non-interrupted GGG-columns have been reported. Assuming a thermodynamically controlled G4 folding, the final conformer will maximize favorable interactions, primarily striving to fill vacant G-core positions for increased stacking interactions but also through additional interactions involving intervening and flanking segments. Depending on the primary sequence, there are various possibilities for intramolecular G insertions into unoccupied G-core positions, leading to distinct structural features with bulged or interrupted G-columns. These approaches are schematically depicted in Fig. 9. In the following, corresponding G4 folds are reviewed with particular emphasis on non-modified quadruplexes whose folding pathway is not guided by conformational preferences of incorporated nucleoside analogs (for the latter, see ref. 65).65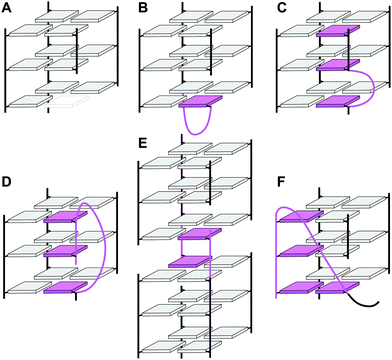 | ||
| Fig. 9 Strategies to fill vacant positions within a quadruplex G-core. (A) Quadruplex with a vacant site, (B) snapback-loop, (C) bulge, (D) D-shaped loop, (E) interlocked G4, (F) V-shaped loop. | ||
G-quadruplexes with a guanine vacancy (vG4)
Deviating from the consensus sequence of a canonical G-quadruplex, sequences with three GGG-tracts and one guanine-deficient GG-tract can assemble into a three-layered quadruplex structure with one tetrad bearing a vacant site. Notably, bioinformatics studies have shown that such sequences are abundant in genomes and may be evolutionarily selected in genes with unique distribution patterns in both eukaryotic and prokaryotic organisms.66,67 The vacant site can easily accept a guanine base from guanine-containing metabolites such as GTP or GMP to form an intact and strongly stabilized G-core, demonstrated to effectively alter DNA replication in vitro at physiological GTP concentration.66 Because G-quadruplexes with guanine vacancies (vG4) are distinct from canonical G4 structures in being able to sense intracellular concentrations of guanine derivates, they have been proposed to play a critical role in gene regulation.On the other hand, the abundance of vG4 forming sequences in the human genome offers a great potential for therapeutic interventions by more specific, high-affinity targeting. Thus, a bifunctional G4-binding peptide guided through a covalently linked guanine base was shown to feature promising selectivity and affinity toward the G-deficient quadruplex associated with strong suppression of in vitro replication.68 From an analytical viewpoint, sensors for guanine derivatives based on quadruplexes with a vacant site have been shown to confer exceptional selectivity toward the analyte.69
Despite the presence of a destabilizing additional thymine bulge in the short and non-contiguous GG-column at its 5′-end, the sequence d[TTGTG(TGGG)3T] containing (12-1) guanines was shown by NMR to fold into a G-deficient intramolecular quadruplex with two G-tetrads and one outer G-triad in a parallel-stranded conformation (Fig. 10A).70 In fact, molecular dynamics simulations established the formation of a G-triad-water complex with water molecules occupying the vacant site in the G-triad plane. Again, the vacancy being a G-binding hotspot can be specifically recognized by external guanine bases. High-affinity binding was observed for linear and cyclic d(AG) and cGAMP dinucleotides when targeting a T deletion mutant d[TTGG(TGGG)3T] lacking the bulge.67
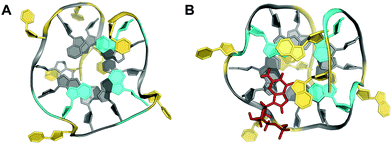 | ||
| Fig. 10 Top view on (A) the vG4 NMR structure formed in K+ solution (10 mM KPi + 35 mM KCl, pH 7.0) from the sequence d[TTGTG(TGGG)3T] with G-triad (colored cyan) stacked on a G-tetrad (PDB 2N60).70 (B) Top view on a dGMP-complexed vG4 structure formed in K+ solution (12.5 mM KPi + 37.5 mM KCl, pH 7.0) from the PDGFR-β gene promoter sequence (PDB 6V0L);71 dGMP (stick model in red) fills the vacant site of the 5′-outer G-layer (colored cyan). Other all-anti G-tetrad core residues are colored grey; loop and flanking residues, yellow. | ||
Another example of a structurally characterized G-deficient quadruplex involves a modified human PDGFR-β gene promoter sequence d(AAG3AG3CG2CG3ACA) termed Pu19m2.71 It was shown to adopt two stable G4 structures formed by the G2-tract shifted toward the 5′- or 3′-terminal quadruplex face with a corresponding vacancy in an outer plane adjacent to either the 3′- or 5′-terminus. The triad layer of the vG4 can again be complemented by the selective and strong external binding of physiologically relevant guanine metabolites such as dGMP, GMP, and cGMP but also by guanine-based drugs (Fig. 10B). Interestingly, metabolite binding is able to modulate the equilibrium between the two G2-shifted isomers, mostly favoring binding to the G-deficient 5′-triad.
Snapback loop G-quadruplexes
If the Pu19m2 sequence of the PDGFR-β promoter with its vG4 fold is extended to also include the wild-type 3′-terminus with another G3-tract, the resulting sequence d(AAG3AG3CG2CG3GCAGGG) designated Pu22m1 was found to adopt a parallel-stranded intramolecular quadruplex with three 1 nt propeller loops and an additional 5 nt lateral loop.39 Here, it is a terminal 3′-G in a syn conformation that intramolecularly fills the vacant site of the third G2-run through a snapback loop structure. Interestingly, the sequence itself features four contiguous runs with ≥3 guanines, expected to fold into a regular three-layered quadruplex without broken strand but with longer second and third loops. Apparently, the high stability of a parallel quadruplex with 1 nt propeller loops outweighs penalties expected for a fourth snapback lateral loop.A snapback approach in combination with a 5′-terminal hairpin structure was also shown to fill a single vacancy left by a short G2-tract. Here, the vacant site acts as an anchor point for the duplex stem-loop in fixing the 5′-terminal G in a syn conformation to the tetrad facing the duplex domain.38
Snapback loops can also bridge distal corners as exemplified by a c-myc promoter sequence d(TGAG3TG4AG3TG4AAG2) containing five guanine tracts. Although able to fold into a regular parallel G4 with 1 and 2 nt propeller loops, it was shown to favor folding into a parallel-stranded fold-back G-quadruplex with the 3′-terminal guanine base filling an empty guanine position within the 3′-tetrad through a diagonal snapback loop.72 The three-dimensional NMR structure of a G10I mutant termed Pu24I demonstrates its parallel fold with 1 nt, 3 nt, and 1 nt propeller loops and a fourth diagonal loop bridging two opposite corners of the 3′-G-tetrad with its terminal syn-G complementing the second G-column (Fig. 11). Single base substitutions suggest that a G·G·A triad within the diagonal loop capping the outer G-tetrad seems a critical structural motif for snapback loop formation in Pu24I. Correspondingly, a stacked G·G·A triad from residues of the diagonal snapback loop was likewise found to stabilize one of the two major G4 conformers formed by a G-rich sequence in the KRAS nuclease hypersensitive element (NHE) region.36 It should be noted, that the addition of further non-G residues at the 3′-terminus may still allow for a fold-back topology but is expected to compromise the thermodynamic stability as suggested by calorimetric studies on mutated 3′-T extended c-myc promoter sequences with five guanine tracts.73
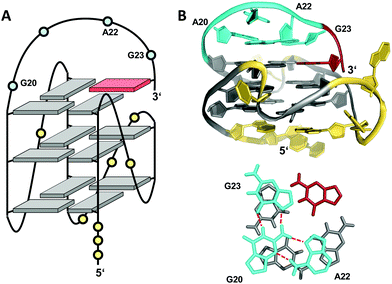 | ||
| Fig. 11 (A) Schematic representation and (B) three-dimensional structure of Pu24I (PDB 2A5P) formed in K+ solution (20 mM KPi + 70 mM KCl, pH 7.0) with residues of the diagonal snapback loop forming a G·G·A triad stacked on the 3′-outer tetrad (bottom);72anti- and syn-guanosines of the G-tetrad core are colored grey and red, respectively; loop and flanking residues, yellow; residues forming the snapback loop, cyan. | ||
Like the extended PDGFR-β promoter sequence, a G-rich c-kit promoter sequence d(AG3AG3CGCTG3AG2AG3) encompassing four G3-tracts and thus capable of forming a regular quadruplex with four continuous G-columns folds into a topology with a snapback loop in K+ solution (Fig. 12).74,75 Again, the pronounced stability of short propeller-type loops in a parallel topology is suggested to drive folding but base pairing alignments in the loops provide for additional stabilization of this structure with several unique features. Here, isolated G10 is recruited to occupy a single outer G-core position and the corresponding G-column is complemented by insertion of the two 3′-terminal anti-G residues aligned in a parallel orientation. The two-residue loop directly following G10 links neighboring corners of the same tetrad. By laterally connecting a broken and continuous G-column of the same strand polarity, it shares features of both propeller and edge-wise loops. The 5 nt snapback connection that follows the fourth G3-column to fill the two vacant sites of the third G-tract with parallel-oriented anti-G residues shows base pairing alignments. It is unusual in spanning two G-quartets with a 3′-flanking G being part of the central tetrad, allowing unrestricted DNA sequence extensions at the 3′-terminus. Formally, this rather long 5 nt loop may also be viewed as a propeller-type loop progressing against the right-handed helicity of the G-core. The overall topology is also conserved when replacing this loop by a hairpin motif within a closely related c-kit based sequence to form a unique quadruplex–duplex junction.38 Clearly, it would also be conceivable to fill the two vacant positions through a conventional lateral-type snapback loop with two terminal syn-Gs in anti-parallel orientation. However, such a conformer may be disfavored by a less stable syn-syn stacking.
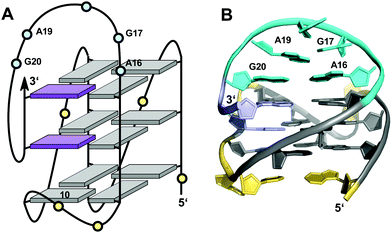 | ||
| Fig. 12 (A) Schematic representation and (B) solution structure of a quadruplex with a distinct type of snapback loop formed by a c-kit promoter sequence in K+ solution (20 mM KPi + 70 mM KCl, pH 7.0; PDB 2O3M);74,75anti-guanosines of the G-core and two inserted 3′-terminal Gs are colored grey and lilac, respectively; loop and flanking residues, yellow; residues of the snapback loop forming A16·G20 and G17·A19 base pairs, cyan. | ||
Quadruplexes with a bulge
Non-consecutive guanosines may assemble into G-quadruplexes that feature a G-column of guanines with interdispersed bases protruding outward to form a bulge. Thus, whereas loop residues connect different columns of the G-tetrad core, a bulge connects adjacent guanines along the same column. Bulges have initially been shown in crystal structures of RNA quadruplexes but more recent reports on several three-dimensional structures of bulged G-quadruplexes in solution attest to their potential prevalence and stability under cellular conditions.36,57,76–83 Consequently, participation of isolated guanines within interrupted G-tracts in the formation of a stable ‘bulged’ G-tetrad core will significantly expand the number of genomic sequences with a potential for G-quadruplex formation.The impact of bulges differing in sequence, size, position, or number on G-quadruplex formation was systematically studied by Mukundan and Phan.80 Their results suggest that bulges can be located at any position in a G-quadruplex structure. However, the stability of quadruplexes with a bulge decreases with increasing bulge size in analogy to the length dependence of propeller loops. Also, the G4 stability will depend on their location but also on the sequence context and the G-quadruplex topology. In general, bulges are destabilizing, limiting the number of individual bulges n compatible with formation of three-layered quadruplexes to n ≤ 3. Destabilization can be attributed to a strained backbone but mostly to the unfavorable entropy of solvating the protruding residues. Thus, entropic effects likely determine a stacking interaction of a thymine bulge with a single-nucleotide propeller loop adenine to reduce the hydrophobic surface area in the long terminal repeat sequence LTR-IV of the proviral HIV-1 genome (Fig. 13A).82 Such rather subtle interactions may in fact explain the different impact of bulges on the thermal stability depending on their position in various topologies.
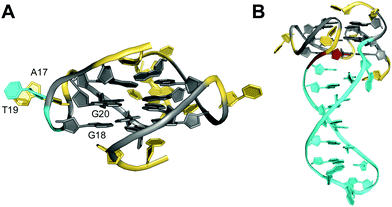 | ||
| Fig. 13 Solution structure of G-quadruplexes with a bulge. (A) Parallel-stranded LTR-IV G-quadruplex from the HIV-1 genome formed in K+ solution (20 mM KPi + 70 mM KCl, pH 7.0; Tm = 50.5 °C; PDB 2N4Y);82anti-G20 following the bulge adopts a north sugar conformation and the bulged T19 stacks onto A17 of the neighboring propeller loop. (B) Parallel-stranded G4 with a bulge forming a stem-loop duplex in K+ solution (20 mM KPi + 30 mM KCl, pH 7.0; Tm = 46.2 °C; PDB 7CLS);85 the G residue following the hairpin-forming bulge adopts a syn conformation. anti- and syn-guanosines of the G-tetrad core are colored grey and red, respectively; loop and flanking residues, yellow; residues in bulges, cyan. | ||
In general, bulges do hardly perturb the G4 core structure which essentially occupies the same conformational space as found for canonical G-quadruplexes. However, guanosine residues adjacent to bulges have been reported to frequently populate an additional range of backbone torsion angles.78 Also, revisiting available quadruplex structures, G-core residues preceding or following the bulge are often found to adopt sugar conformations in the north rather than in the more typical south domain. It should be noted, however, that in most cases no restraints for sugar dihedral angles were employed for generating the NMR solution structures.
A unique 2 nt GA bulge in a G-quadruplex formed by a G-rich sequence in the regulatory region of a RANKL gene connects anti- and syn-guanosines that occupy G-core positions of a parallel G4.84 Consequently, in order to maintain formation and proper stacking of the G-tetrads, the bulge must provide for a turn of the backbone in adopting a pseudo-loop conformation. Remarkably, the corresponding G4-forming sequence encompasses four G3-tracts to allow for a regular three-layered quadruplex. Assuming the bulge to be destabilizing, the bulge-containing fold seems to be driven by a shorter overall 1-3-1 when compared to a 1-3-3 propeller loop architecture as expected for a bulge-free parallel topology.
Recently, base-paired duplex bulges of different size were incorporated into various positions of a G-quadruplex scaffold, demonstrating their noticeable stabilization when compared to unstructured bulges.85 In fact, thermal stabilities of duplex bulges are slightly increased with increasing bulge sizes, following a similar trend as observed for G4 hairpin loops. The formed quadruplex–duplex junction is reminiscent of an orthogonally aligned propeller-type stem-loop structure with a first disrupted base pair to allow for a progressive transition from the quadruplex to the duplex segment associated with an increase in strand separation (Fig. 13B). However, in contrast to a propeller-type hairpin loop the double-helical foldback bulge continuously stacks onto the 3′-outer G-tetrad and only the first unpaired base projects outward from the groove.
D-Shaped loop
Unlike bulges that link two split G residues within the same G-column in a consecutive way, another distinct type of loop connects residues of a column located at opposite faces of the G-core. Owing to its characteristic progression it has also been termed D-shaped loop.26 This peculiar arrangement positions the 5′- or 3′-terminal G of a d(G2NxG) or d(GNxG2) tract between the other two G residues when forming a G-quadruplex column (see Fig. 9D). In this regard it is reminiscent of a structural motif reported for a short fragment of telomeric DNA from S. cerevisiae. Here, an unusual pseudo-circular G-hairpin with a compact core of three GG base pairs is formed and a chain reversal within a continuous G3-tract places the 3′-terminal G between the two preceding G residues in the base-paired structure.86In a G-quadruplex, such a structural motif was shown for a G-rich VEGF aptamer carrying three locked nucleic acid modifications.26 Here, a 2 nt D-shaped loop fills a vacant position within the same column by bridging two corners on opposite sides of the G-core (Fig. 14). Notably, with all three tetrads featuring the same polarity and all core guanosines adopting an anti conformation as demonstrated by NMR data analysis, there seems to be no strand inversion between the flanking outer G-core residues as would be expected for this structural motif. Interestingly, however, an easy switch to a syn conformation was observed for the 3′-flanking G during structure calculations. A 0 nt propeller loop bridging two tetrad planes precedes and another 2 nt loop directly follows the V-shaped loop. The unusual 2 nt loop ties two parallel-oriented G positions at adjacent corners of the same tetrad in analogy to a corresponding loop in the c-kit promoter G-quadruplex.74,75
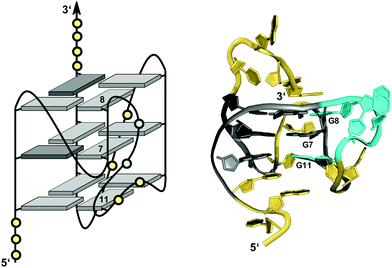 | ||
| Fig. 14 Schematic representation (left) and three-dimensional solution structure (right) of a G-quadruplex with a D-shaped loop derived from a VEGF aptamer with locked LNAG residues in K+ solution (10 mM KPi + 40 mM KCl, pH 7.0; Tm = 52 °C; PDB 2M53);26anti-guanosines and LNA analogs of the G-core are colored in light and dark grey, respectively; loop and flanking residues, yellow; residues of D-shaped loop, cyan. | ||
Another example for a D-shaped loop comes from a guanine-rich 36 nt RNA motif named sc1 capable of quadruplex formation. The solution structure of the sc1 RNA complexed with an arginine-glycine-rich RGG peptide from the FMRP protein reveals a G-quadruplex domain connected to a flanking duplex stem.87 The three-layered all-anti G4 is composed of two stacked tetrads of the same polarity and an additional G-tetrad of opposite polarity facing the duplex domain. Here, a strand polarity inversion within one G3-tract and two 1 nt D-shaped loops associated with a flipped backbone connect the inverted G-tetrad with the other two G-tetrad layers.
Interlocked structures
Interlocked structures are composed of more than one separate G-rich strand and in the past have often been associated with the formation of G-wires. The latter can form if G-rich strands associate out-of-register to present ‘sticky’ ends. Two such slipped structures may subsequently dimerize through their terminal free G residues to form an extra G-tetrad. Thus, d(GGGT) may align into an octameric complex with five stacked G-tetrads in addition to the tetramolecular d(GGGT)4 with in-register strand association (Fig. 15A).88 If association is enabled at both 5′- and 3′-ends, self-assembly can lead to large nanostructures by the growth of an interlocked G4 in both directions.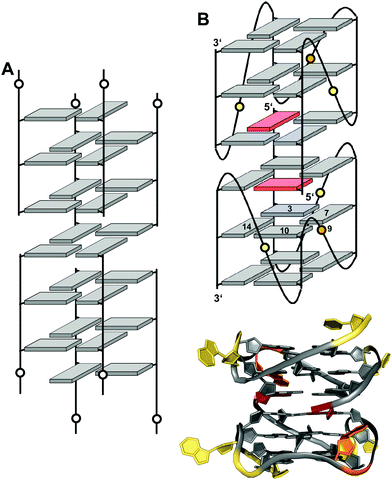 | ||
| Fig. 15 Interlocked G-quadruplexes. (A) Schematic representation of two interlocked d(GGGT)4 out-of-register quadruplexes forming an octameric species.88 (B) Schematic representation (top) and three-dimensional structure (bottom) of an interlocked quadruplex formed in K+ solution (90 mM KCl, pH 7.0) by an HIV-integrase aptamer (PDB 1Y8D);98 the pair of G-tetrads at the dimer interface are mutually filled with a 5′-terminal syn-G1 from the other strand; anti- and syn-guanosines of the G-tetrad core are colored grey and red, respectively; loop and flanking residues, yellow; loop adenines A9 aligned in-plane of the tetrad to form a pentad, orange. | ||
A first model of a G-wire formed by the telomeric DNA oligonucleotide d(G4T2G4) was proposed more than 25 years ago89,90 but its structural diversity could only be demonstrated by atomic force microscopy in the recent past.91 Expanding on the self-associative potential of G-rich sequences in a slipped alignment, oligomerization was also shown to be supported by GC overhangs that serve as cohesive ‘sticky’ ends to form two interfacial GCGC-tetrads by hydrogen bonding through Watson–Crick and Hoogsteen guanine edges.92–94 In G-wires, self-recognition and self-assembly relies on G-quartet formation and is expected to be promoted by hydrophobic effects but also by the enthalpic gain of multiple G-tetrad stacking. Correspondingly, these interlocked structures show high thermal stabilities but due to their slow kinetics of formation their population and length strongly depends on concentration, temperature, and cations present. Of note, some of the higher-order G4 structures suggested to coexist in particular with parallel-stranded G-quadruplexes23,95,96 may possibly also be traced to the formation of such high-melting interlocked structures.
Narrowing the definition of interlocked G-quadruplexes, G-tetrads at the interface between two G-deficient quadruplex folds may be mutually completed by Gs from the other subunit. This enables the sequence d(G3AG2T3G3AT), bearing only three G-tracts, to fold into a dimeric four-layered quadruplex.97 Each monomer adopts a compact domain with a 1 nt propeller loop, a 3 nt lateral loop, and a 0 nt V-shaped loop (see below). The dimeric interface features a pair of stacked A·(G·G·G·G) pentads through the interaction and in-plane alignment of the propeller loop adenine with a G-quartet. Also, each pentad is completed through the insertion of a 5′-terminal syn-G residue of the other monomer. The pentad stacks upon a tetrad in each monomer supported by one broken and three continuous GG-columns.
Likewise, the 93del aptamer d(GGGGTGGGAGGAGGGT), an HIV-1 integrase inhibitor, forms a very stable six-layered G-quadruplex interlocked dimer in K+ solution (Fig. 15B).98 Each monomeric subunit contains one A·(G·G·G·G) pentad sandwiched between two G-tetrads with the G-tetrad at the interlocking interface complemented by the 5′-terminal syn-G from the first G4-tract of the other monomer. All G-columns within each monomer are parallel and linked by two 1 nt propeller loops bridging three G-tetrad layers. A second adenosine propeller loop that bridges two G-tetrad layers participates in A·(G·G·G·G) pentad formation.
Based on the 93del aptamer, sequence variants forming corresponding interlocked G-quadruplex dimers were rationally designed. These encompass a first long G4-tract to compensate with its 5′-G for a G-deficient tetrad of the other monomer, two medium G3-tracts, and another short G2-tract being positioned as second, third, or fourth G-run.99 Indeed, very stable interlocked quadruplexes were demonstrated to form and may constitute robust scaffolds for technological applications.
Expanding on the architecture of locked quadruplexes, a unique intra-locked G4 structure was recently reported for the 28mer G-rich sequence d[(TGG)4TTG(TGG)3TTGT] harboring multiple G2-tracts and two single G residues.100 This sequence was shown to fold into a structure with two stacked bi-layered subunits formed by its 5′- and 3′-domains. Additionally, intramolecular locking is achieved by the incorporation of a guanine from the 5′-subunit into the G-deficient interfacial G-layer of the 3′-subunit.
V-loop quadruplexes
V-loops are one of the most prominent non-canonical structural elements in G-quadruplexes and have received growing attention in recent years. V-shaped loops connect two adjacent G-columns by bridging two or three G-tetrad layers (see Fig. 9F). However, in contrast to a propeller-type connection one of the G-columns is broken and the two linked G-tracts are generally oriented anti-parallel to each other. The formation of 0 nt V-shaped loops is most common, but in some cases 1 nt or 2 nt V-loops have also been reported.54,100,101V-loops can exhibit high intrinsic stability and may even compete with canonical topologies. Thus, modifying all matching anti-G positions with strongly anti-favoring LNAG analogs in the telomeric sequence d(G4T4G4) from Oxytricha nova resulted in a rearrangement of the bimolecular anti-parallel quadruplex into a unique scaffold with a topology termed V4 fold.102 Here, all four G-stretches within two strands fold back in a V-shaped loop with an LNA residue at their 3′-end and interact with each of the other three G-stretches through the formation of four G-tetrads. The V-loop 5′-anchoring position is generally occupied by a syn-G being part of a discontinuous G-column. In order to trace favorable and unfavorable contributions to V-loop formation, various sugar-modified G analogs have recently been introduced at specific positions of a (3+1) hybrid quadruplex, triggering rearrangements into a V-loop structure.103–105 Detailed analysis of dual-modified V-loop quadruplexes bearing different combinations of G-analogs demonstrated that often overlooked sugar conformational preferences rather than glycosidic conformations were major contributors to V-loop stability. Thus, a stable V-loop structure was even formed when inserting LNAG with its fixed C3′-endo conformation (north) at the V-loop 5′-anchoring site to enforce a strongly disfavored syn conformation when followed by another 3′-flanking anti-LNAG.105 On the other hand, a sugar pucker in the north domain for both 5′- and 3′-flanking residues seems to match backbone conformational requirements of a conventional 0 nt V-shaped loop (Fig. 16A, top). In fact, a corresponding 5′-(syn,north)-(anti,north)-3′ conformation for V-loop flanking residues is likewise found for other V-shaped loops in unmodified quadruplexes and represents a characteristic feature for such conventional loops.106–108 Owing to the syn and anti anchor residues participating in G-tetrads of reversed polarity, there is no apparent strand polarity inversion inherent to the V-loop but rather between the 3′-flanking G and the following G within the same G-tract. Interestingly, a sharp turn of the sugar-phosphate backbone at the inversion site and a north-type sugar pucker of the 3′-anchoring residue places its O4′ and O5′ oxygen atoms in close vicinity to H8 of the following G to also allow for corresponding C–H⋯O interactions (Fig. 16A, bottom).
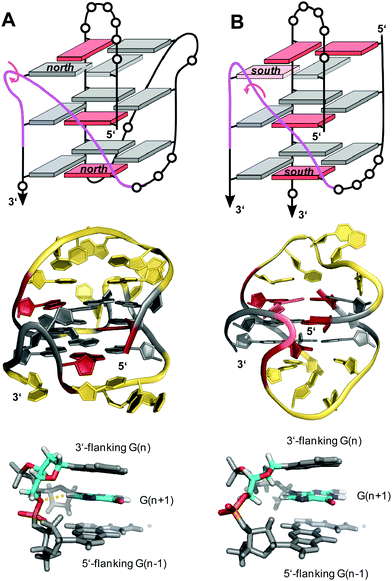 | ||
| Fig. 16 V-loop topology with polarity inversion sites indicated by circular arrows (top), solution structure (center), and backbone conformation for the V-loop (bottom) comprising a 3 nt stretch that is colored magenta in the topological representations on top. (A) Monomolecular G4 formed in K+ solution (20 mM KPi + 80 mM KCl, pH 6.8; Tm = 54.5 °C) with conventional V-loop and O5′(n)-H8(n + 1) interactions (PDB 5ZEV).106 (B) Bimolecular G4 in K+ solution (15 mM KCl, pH 5.5) with alternative V-loop and solvent-exposed phosphate (PDB 1U64).109 G-core guanosines in anti, syn, and ‘low-syn’ conformation are colored grey, red, and light red, respectively. | ||
A second type of V-loop conformation has originally been suggested based on a sequence bearing a south/south-east-favoring 2′-fluoro-arabinoguanosine analog at the 3′-flanking position.104,105 In these alternative V-loop conformers, south-puckered residues are mostly found for both V-loop framing positions, allowing to differentiate V-shaped loops according to two distinct sugar conformational preferences (Fig. 16B, top). Whereas a syn conformation at the 5′-anchor site seems mandatory for all regular V-loops, glycosidic torsion angles at the 3′-end of the alternative V-loop are typically outside the anti/high-anti range and rather adopt torsion angles in a less defined ‘low-syn’ range.41,109,110 As a consequence, a sugar-phosphate backbone inversion can formally be localized within the V-loop in this case. Also, larger inter-atomic distances with a more solvent-exposed phosphate of the 3′-flanking residue prevent C–H⋯O pseudo-hydrogen bond contacts between the latter and the subsequent G as observed for a conventional type of V-loop, with possible implications for other intermolecular interactions (Fig. 16B, bottom).
Due to a lessening of conformational restraints exerted by the V-shaped loop, few quadruplexes with a less compact architecture, e.g., with more flexible 1 nt or 2 nt V-loops or with neighboring bulges adjacent to the loop, may feature conformers located slightly outside of either of the two characteristic conformational clusters.100,101 Of note, foldback bulges in two recently reported parallel-stranded quadruplexes enforce a single syn conformer for the 3′-linked G of a central discontinuous G-tract (see above and Fig. 13B).84,85 As a result, the loop following this G-core residue with its inverted backbone orientation may be viewed as a 1 nt V-shaped loop rather than a regular propeller-type loop.
Finally, a unique two-layered anti-parallel quadruplex derived from the AGRO100 aptamer combines a 1 nt V-shaped loop with a 3′-terminal domain forming characteristic base pairing alignments.54 The 3′-peripheral motif progresses along two sharp U-turns to form two additional layers composed of a T·T·G triad and a G·T base pair capping the 5′-outer tetrad (Fig. 17). It is attached by a non-terminal guanosine to the G4 core, filling a vacant G4 position in a snapback-type arrangement. The compact structural domain of the peripheral sequence was shown to possibly serve as a modular unit, able to replace a diagonal snapback loop in other G4 structures. Notably, in contrast to most V-loop structures the unusual 1 nt V-loop spanning two tetrad planes features a 5′-anchoring guanosine in anti conformation.
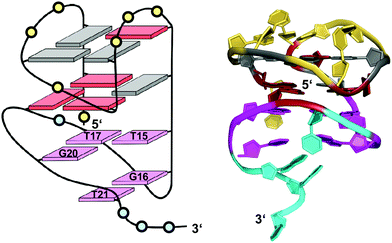 | ||
| Fig. 17 Schematic representation and three-dimensional solution structure for a V-loop quadruplex of the AGRO100 derived sequence d(TG2TTGTG2TTTG2TGTTG2TG2T) in K+ solution (20 mM KPi + 70 mM KCl, pH 7.0; Tm = 39 °C; PDB 6JCD);54anti- and syn-guanosines of the G-tetrad core are colored grey and red, respectively; loop and flanking residues of the G4 core structure and of the 3′-peripheral motif are colored yellow and cyan, respectively; T·T·G triad and G·T base pair, magenta. | ||
Summary and perspectives
G-rich sequences covering a wide range of G-runs of different number and length have been demonstrated to fold into four-stranded G-quadruplexes. As surveyed in this review, G-deficiencies as a result of short G-tracts may be compensated by unique structural motifs supplementing vacant sites. These include V- and D-shaped loops, snapback loops, bulges, and interlocked species. On the other hand, additional stacking interactions through G4 association but also through capping base triads or base pairs may considerably stabilize a particular G4 architecture. Formation of corresponding motifs will depend on the particular sequence context and on the superposition of additional interactions that are often difficult to anticipate based on the primary sequence. However, these interactions may even be strong enough to induce folding into a non-conventional structure even if the sequence allowed folding into a canonical topology.Based on the rapidly growing number of reported crystallographic and NMR G-quadruplex structures, our knowledge on the structural organization and stabilizing inherent interactions of unusual structural motifs has considerably grown. Thus, we are beginning to recognize and understand major contributors to particular topological features. These include tertiary interactions between different domains of the folded quadruplex to form triads, base pairs, or non-canonical tetrads but also more subtle conformational preferences of individual residues with their often decisive impact on equilibria between G4 conformers being close in energy. Sequences that fold into non-canonical G4 structures featuring interrupted G-columns have already been successfully designed.38,99,111 However, whereas our ability to decipher the code that relates a G-rich oligonucleotide sequence with four closely spaced G3-tracts to a preferred G4 topology increases, understanding and predicting the folding of irregular G-rich sequences still poses a challenge.
Detailed insight into folding pathways and into interactions enforcing particular structural motifs will be important for the identification of G-rich fragments prone to G-quadruplex formation and also for a successful engineering of quadruplex architectures for various technology-based G4 applications. On the other hand, non-canonical G4 structures offer additional opportunities in their specific targeting for both pharmaceutical and technological purposes. The majority of known G4 ligands binds through stacking interactions onto a G-tetrad. Also, attempts to increase selectivity with less off-target effects based on different groove dimensions or loop conformations has only brought limited success to-date. Exploiting various non-conventional structural motifs may in fact expand our toolbox for achieving more selectivity. G-deficient quadruplexes with a vacant site constitute promising targets for a specific and high-affinity binding of bifunctional ligands that are guided by a covalently linked guanine base. Also, interrupted G-tracts with their opening may potentially support insertion of planar ligands between tetrad planes of the quadruplex. Finally, quadruplex–duplex junctions are expected to be widespread in a cellular environment either through hairpin-type loop domains within the G4 architecture or through G-quadruplexes extruding from a B-type duplex as anticipated for oncogenic promoter sequences. Such interfaces between different structural domains have attracted growing interest in recent years and may provide for unique target sites for G4 drugs.112–114 In fact, binding to quadruplex–duplex junctions by appropriate ligands has already been shown to be guided by strong interactions, making junctions one of several promising target sites for the future design of more efficient G4-binding drugs.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Deutsche Forschungsgemeinschaft for financial support (grant no. WE 1933/15-1).Notes and references
- E. Y. N. Lam, D. Beraldi, D. Tannahill and S. Balasubramanian, Nat. Commun., 2013, 4, 1796 CrossRef.
- A. Laguerre, K. Hukezalie, P. Winckler, F. Katranji, G. Chanteloup, M. Pirrotta, J.-M. Perrier-Cornet, J. M. Y. Wong and D. Monchaud, J. Am. Chem. Soc., 2015, 137, 8521–8525 CrossRef CAS.
- R. Hänsel-Hertsch, M. D. Antonio and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2017, 18, 279–284 CrossRef.
- S. Neidle, Nat. Rev. Chem., 2017, 1, 0041 CrossRef CAS.
- I. Russo Krauss, A. Merlino, C. Giancola, A. Randazzo, L. Mazzarella and F. Sica, Nucleic Acids Res., 2011, 39, 7858–7867 CrossRef CAS.
- V. R. de Soultrait, P.-Y. Lozach, R. Altmeyer, L. Tarrago-Litvak, S. Litvak and M. L. Andréola, J. Mol. Biol., 2002, 324, 195–203 CrossRef CAS.
- A.-M. Chiorcea-Paquim, R. Eritja and A. M. Oliveira-Brett, J. Nucleic Acids, 2018, 5307106 Search PubMed.
- J. L. Neo, K. Kamaladasan and M. Uttamchandani, Curr. Pharm. Des., 2012, 18, 2048–2057 CrossRef CAS.
- N. Borovok, N. Iram, D. Zikich, J. Ghabboun, G. I. Livshits, D. Porath and A. B. Kotlyar, Nucleic Acids Res., 2008, 36, 5050–5060 CrossRef CAS.
- J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2005, 33, 2908–2916 CrossRef CAS.
- A. K. Todd, M. Johnston and S. Neidle, Nucleic Acids Res., 2005, 33, 2901–2907 CrossRef CAS.
- A. B. Sahakyan, V. S. Chambers, G. Marsico, T. Santner, M. D. Antonio and S. Balasubramanian, Sci. Rep., 2017, 7, 14535 CrossRef.
- E. Puig Lombardi and A. Londoño-Vallejo, Nucleic Acids Res., 2020, 48, 1–15 CrossRef.
- D. Bhattacharyya, G. M. Arachchilage and S. Basu, Front. Chem., 2016, 4, 38 Search PubMed.
- M. Webba da Silva, Chem. – Eur. J, 2007, 13, 9738–9745 CrossRef CAS.
- F. Fogolari, H. Haridas, A. Corazza, P. Viglino, D. Corà, M. Caselle, G. Esposito and L. E. Xodo, BMC Struct. Biol., 2009, 9, 64 CrossRef.
- S. A. Dvorkin, A. I. Karsisiotis and M. Webba da Silva, Sci. Adv., 2018, 4, eaat3007 CrossRef CAS.
- B. Karg, L. Haase, A. Funke, J. Dickerhoff and K. Weisz, Biochemistry, 2016, 55, 6949–6955 CrossRef CAS.
- X. Tong, W. Lan, X. Zhang, H. Wu, M. Liu and C. Cao, Nucleic Acids Res., 2011, 39, 6753–6763 CrossRef CAS.
- X. Cang, J. Šponer and T. E. Cheatham, Nucleic Acids Res., 2011, 39, 4499–4512 CrossRef CAS.
- J. Šponer, A. Mládek, N. Špačková, X. Cang, T. E. Cheatham and S. Grimme, J. Am. Chem. Soc., 2013, 135, 9785–9796 CrossRef.
- P. Hazel, J. Huppert, S. Balasubramanian and S. Neidle, J. Am. Chem. Soc., 2004, 126, 16405–16415 CrossRef CAS.
- N. Smargiasso, F. Rosu, W. Hsia, P. Colson, E. S. Baker, M. T. Bowers, E. De Pauw and V. Gabelica, J. Am. Chem. Soc., 2008, 130, 10208–10216 CrossRef CAS.
- M. Cheng, Y. Cheng, J. Hao, G. Jia, J. Zhou, J. L. Mergny and C. Li, Nucleic Acids Res., 2018, 46, 9264–9275 CrossRef CAS.
- A. Piazza, X. Cui, M. Adrian, F. Samazan, B. Heddi, A. T. Phan and A. G. Nicolas, eLife, 2017, 6, e26884 CrossRef.
- M. Marušič, R. N. Veedu, J. Wengel and J. Plavec, Nucleic Acids Res., 2013, 41, 9524–9536 CrossRef.
- T. Fujii, P. Podbevšek, J. Plavec and N. Sugimoto, J. Inorg. Biochem., 2017, 166, 190–198 CrossRef CAS.
- V. Kuryavyi, A. T. Phan and D. J. Patel, Nucleic Acids Res., 2010, 38, 6757–6773 CrossRef CAS.
- M. Trajkovski, M. Webba da Silva and J. Plavec, J. Am. Chem. Soc., 2012, 134, 4132–4141 CrossRef CAS.
- M. Webba da Silva, M. Trajkovski, Y. Sannohe, N. Ma’ani Hessari, H. Sugiyama and J. Plavec, Angew. Chem., Int. Ed., 2009, 48, 9167–9170 CrossRef CAS.
- B. Karg, S. Mohr and K. Weisz, Angew. Chem., Int. Ed., 2019, 58, 11068–11071 CrossRef CAS.
- A. Bugaut and S. Balasubramanian, Biochemistry, 2008, 47, 689–697 CrossRef CAS.
- P. Hazel, J. Huppert, S. Balasubramanian and S. Neidle, J. Am. Chem. Soc., 2004, 126, 16405–16415 CrossRef CAS.
- A. Guédin, J. Gros, P. Alberti and J. L. Mergny, Nucleic Acids Res., 2010, 38, 7858–7868 CrossRef.
- S. Amrane, M. Adrian, B. Heddi, A. Serero, A. Nicolas, J. L. Mergny and A. T. Phan, J. Am. Chem. Soc., 2012, 134, 5807–5816 CrossRef CAS.
- J. Marquevielle, C. Robert, O. Lagrabette, M. Wahid, A. Bourdoncle, L. E. Xodo, J.-L. Mergny and G. F. Salgado, Nucleic Acids Res., 2020, 48, 9336–9345 CrossRef.
- D. J. E. Yue, K. W. Lim and A. T. Phan, J. Am. Chem. Soc., 2011, 133, 11462–11465 CrossRef CAS.
- K. W. Lim and A. T. Phan, Angew. Chem., Int. Ed., 2013, 52, 8566–8569 CrossRef CAS.
- Y. Chen, P. Agrawal, R. V. Brown, E. Hatzakis, L. Hurley and D. Yang, J. Am. Chem. Soc., 2012, 134, 13220–13223 CrossRef CAS.
- K. W. Lim, Z. J. Khong and A. T. Phan, Biochemistry, 2014, 53, 247–257 CrossRef CAS.
- E. Butovskaya, B. Heddi, B. Bakalar, S. N. Richter and A. T. Phan, J. Am. Chem. Soc., 2018, 140, 13654–13662 CrossRef CAS.
- R. Jin, B. L. Gaffney, C. Wang, R. A. Jones and K. J. Breslauer, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 8832–8836 CrossRef CAS.
- L. Petraccone, E. Erra, V. Esposito, A. Randazzo, L. Mayol, L. Nasti, G. Barone and C. Giancola, Biochemistry, 2004, 43, 4877–4884 CrossRef CAS.
- R. F. Macaya, P. Schultze, F. W. Smith, J. A. Roe and J. Feigon, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3745–3749 CrossRef CAS.
- I. Smirnov and R. H. Shafer, Biochemistry, 2000, 39, 1462–1468 CrossRef CAS.
- P. Schultze, R. F. Macaya and J. Feigon, J. Mol. Biol., 1994, 235, 1532–1547 CrossRef CAS.
- L. Martino, A. Virno, A. Randazzo, A. Virgilio, V. Esposito, C. Giancola, M. Bucci, G. Cirino and L. Mayol, Nucleic Acids Res., 2006, 34, 6653–6662 CrossRef CAS.
- K. W. Lim, S. Amrane, S. Bouaziz, W. Xu, Y. Mu, D. J. Patel, K. N. Luu and A. T. Phan, J. Am. Chem. Soc., 2009, 131, 4301–4309 CrossRef CAS.
- Z. Zhang, J. Dai, E. Veliath, R. A. Jones and D. Yang, Nucleic Acids Res., 2009, 38, 1009–1021 CrossRef.
- M. Lenarčič Živković, J. Rozman and J. Plavec, Angew. Chem., Int. Ed., 2018, 57, 15395–15399 CrossRef.
- A. Kettani, S. Bouaziz, W. Wang, R. A. Jones and D. J. Patel, Nat. Struct. Biol., 1997, 4, 382–389 CrossRef CAS.
- A. Kettani, G. Basu, A. Gorin, A. Majumdar, E. Skripkin and D. J. Patel, J. Mol. Biol., 2000, 301, 129–146 CrossRef CAS.
- V. Kuryavyi, A. Kettani, W. Wang, R. Jones and D. J. Patel, J. Mol. Biol., 2000, 295, 455–469 CrossRef CAS.
- T. H. A. Truong, F. R. Winnerdy and A. T. Phan, Angew. Chem., Int. Ed., 2019, 58, 13834–13839 CrossRef CAS.
- B. Bakalar, B. Heddi, E. Schmitt, Y. Mechulam and A. T. Phan, Angew. Chem., Int. Ed., 2019, 58, 2331–2335 CrossRef CAS.
- F. R. Winnerdy, B. Bakalar, A. Maity, J. J. Vandana, Y. Mechulam, E. Schmitt and A. T. Phan, Nucleic Acids Res., 2019, 47, 8272–8281 CrossRef CAS.
- N. Q. Do, W. J. Chung, T. H. A. Truong, B. Heddi and A. T. Phan, Nucleic Acids Res., 2017, 45, 7487–7493 CrossRef CAS.
- W. J. Chung, B. Heddi, E. Schmitt, K. W. Lim, Y. Mechulam and A. T. Phan, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2729–2733 CrossRef CAS.
- A. Kettani, A. Gorin, A. Majumdar, T. Hermann, E. Skripkin, H. Zhao, R. Jones and D. J. Patel, J. Mol. Biol., 2000, 297, 627–644 CrossRef CAS.
- Y. Mishima, H. Kaizu and R. Kominami, J. Biol. Chem., 1997, 272, 26578–26584 CrossRef CAS.
- M. Heller, E. Flemington, E. Kieff and P. Deininger, Mol. Cell. Biol., 1985, 5, 457–465 CrossRef CAS.
- A. Matsugami, K. Ouhashi, M. Kanagawa, H. Liu, S. Kanagawa, S. Uesugi and M. Katahira, J. Mol. Biol., 2001, 313, 255–269 CrossRef CAS.
- A. Matsugami, T. Okuizumi, S. Uesugi and M. Katahira, J. Biol. Chem., 2003, 278, 28147–28153 CrossRef CAS.
- N. Q. Do, K. W. Lim, M. H. Teo, B. Heddi and A. T. Phan, Nucleic Acids Res., 2011, 39, 9448–9457 CrossRef CAS.
- L. Haase, B. Karg and K. Weisz, ChemBioChem, 2019, 20, 985–993 CrossRef CAS.
- X. M. Li, K. W. Zheng, J. Y. Zhang, H. H. Liu, Y. De He, B. F. Yuan, Y. H. Hao and Z. Tan, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 14581–14586 CrossRef CAS.
- F. R. Winnerdy, P. Das, B. Heddi and A. T. Phan, J. Am. Chem. Soc., 2019, 141, 18038–18047 CrossRef CAS.
- Y. De, He, K. W. Zheng, C. J. Wen, X. M. Li, J. Y. Gong, Y. H. Hao, Y. Zhao and Z. Tan, J. Am. Chem. Soc., 2020, 142, 11394–11403 CrossRef.
- X. M. Li, K. W. Zheng, Y. H. Hao and Z. Tan, Angew. Chem., Int. Ed., 2016, 55, 13759–13764 CrossRef CAS.
- B. Heddi, N. Martín-Pintado, Z. Serimbetov, T. M. A. Kari and A. T. Phan, Nucleic Acids Res., 2016, 44, 910–916 CrossRef CAS.
- K. B. Wang, J. Dickerhoff, G. Wu and D. Yang, J. Am. Chem. Soc., 2020, 142, 5204–5211 CrossRef CAS.
- A. T. Phan, V. Kuryavyi, H. Y. Gaw and D. J. Patel, Nat. Chem. Biol., 2005, 1, 167–173 CrossRef CAS.
- J. Jana and K. Weisz, Chem. – Eur. J., 2020, 26, 17242–17251 CrossRef CAS.
- A. T. Phan, V. Kuryavyi, S. Burge, S. Neidle and D. J. Patel, J. Am. Chem. Soc., 2007, 129, 4386–4392 CrossRef CAS.
- D. Wei, G. N. Parkinson, A. P. Reszka and S. Neidle, Nucleic Acids Res., 2012, 40, 4691–4700 CrossRef CAS.
- B. Pan, Y. Xiong, K. Shi and M. Sundaralingam, Structure, 2003, 11, 1423–1430 CrossRef CAS.
- B. Pan, K. Shi and M. Sundaralingam, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 3130–3134 CrossRef CAS.
- M. Meier, A. Moya-Torres, N. J. Krahn, M. D. McDougall, G. L. Orriss, E. K. S. McRae, E. P. Booy, K. McEleney, T. R. Patel, S. A. McKenna and J. Stetefeld, Nucleic Acids Res., 2018, 46, 5319–5331 CrossRef CAS.
- V. T. Mukundan, N. Q. Do and A. T. Phan, Nucleic Acids Res., 2011, 39, 8984–8991 CrossRef CAS.
- V. T. Mukundan and A. T. Phan, J. Am. Chem. Soc., 2013, 135, 5017–5028 CrossRef CAS.
- H. Martadinata and A. T. Phan, Biochemistry, 2014, 53, 1595–1600 CrossRef CAS.
- B. De Nicola, C. J. Lech, B. Heddi, S. Regmi, I. Frasson, R. Perrone, S. N. Richter and A. T. Phan, Nucleic Acids Res., 2016, 44, 6442–6451 CrossRef CAS.
- A. Sengar, J. J. Vandana, V. S. Chambers, M. Di Antonio, F. R. Winnerdy, S. Balasubramanian and A. T. Phan, Nucleic Acids Res., 2019, 47, 1564–1572 CrossRef CAS.
- M. Lenarčič Živković, J. Rozman and J. Plavec, Molecules, 2020, 25, 4867 CrossRef.
- T. Q. N. Nguyen, K. W. Lim and A. T. Phan, Nucleic Acids Res., 2020, 48, 10567–10575 CrossRef.
- M. Gajarský, M. Lenarčič Živković, P. Stadlbauer, B. Pagano, R. Fiala, J. Amato, L. Tomáška, J. Šponer, J. Plavec and L. Trantírek, J. Am. Chem. Soc., 2017, 139, 3591–3594 CrossRef.
- A. T. Phan, V. Kuryavyi, J. C. Darnell, A. Serganov, A. Majumdar, S. Ilin, T. Raslin, A. Polonskaia, C. Chen, D. Clain, R. B. Darnell and D. J. Patel, Nat. Struct. Mol. Biol., 2011, 18, 796–804 CrossRef CAS.
- Y. Krishnan-Ghosh, D. Liu and S. Balasubramanian, J. Am. Chem. Soc., 2004, 126, 11009–11016 CrossRef CAS.
- T. C. Marsh and E. Henderson, Biochemistry, 1994, 33, 10718–10724 CrossRef CAS.
- T. C. Marsh, J. Vesenka and E. Henderson, Nucleic Acids Res., 1995, 23, 696–700 CrossRef CAS.
- K. Bose, C. J. Lech, B. Heddi and A. T. Phan, Nat. Commun., 2018, 9, 1959 CrossRef.
- M. Webba da Silva, Biochemistry, 2003, 42, 14356–14365 CrossRef CAS.
- M. Webba da Silva, Biochemistry, 2005, 44, 3754–3764 CrossRef CAS.
- N. Ma’Ani Hessari, L. Spindler, T. Troha, W. C. Lam, I. Drevenšek-Olenik and M. Webba Da Silva, Chem. – Eur. J., 2014, 20, 3626–3630 CrossRef.
- H. T. Le, M. C. Miller, R. Buscaglia, W. L. Dean, P. A. Holt, J. B. Chaires and J. O. Trent, Org. Biomol. Chem., 2012, 10, 9393–9404 RSC.
- V. Rauser and E. Weinhold, ChemBioChem, 2020, 21, 2445–2448 CrossRef CAS.
- N. Zhang, A. Gorin, A. Majumdar, A. Kettani, N. Chernichenko, E. Skripkin and D. J. Patel, J. Mol. Biol., 2001, 311, 1063–1079 CrossRef CAS.
- A. T. Phan, V. Kuryavyi, J.-B. Ma, F. Aurélie, M.-L. Andréola and D. J. Patel, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 634–639 CrossRef CAS.
- A. T. Phan and N. Q. Do, Nucleic Acids Res., 2013, 41, 2683–2688 CrossRef CAS.
- A. Maity, F. R. Winnerdy, W. D. Chang, G. Chen and A. T. Phan, Nucleic Acids Res., 2020, 48, 3315–3327 CrossRef CAS.
- M. Adrian, D. J. Ang, C. J. Lech, B. Heddi, A. Nicolas and A. T. Phan, J. Am. Chem. Soc., 2014, 136, 6297–6305 CrossRef CAS.
- J. T. Nielsen, K. Arar and M. Petersen, Angew. Chem., Int. Ed., 2009, 48, 3099–3103 CrossRef CAS.
- L. Haase, J. Dickerhoff and K. Weisz, Chem. – Eur. J., 2020, 26, 524–533 CrossRef CAS.
- L. Haase and K. Weisz, Chem. Commun., 2020, 56, 4539–4542 RSC.
- L. Haase and K. Weisz, Nucleic Acids Res., 2020, 48, 10555–10566 CrossRef.
- Y. Liu, W. Lan, C. Wang and C. Cao, J. Biol. Chem., 2018, 293, 8947–8955 CrossRef CAS.
- M. Marušič and J. Plavec, Molecules, 2019, 24, 1294 CrossRef.
- V. Kuryavyi and D. J. Patel, Structure, 2010, 18, 73–82 CrossRef CAS.
- M. Črnugelj, P. Šket and J. Plavec, J. Am. Chem. Soc., 2003, 125, 7866–7871 CrossRef.
- C. Wan, W. Fu, H. Jing and N. Zhang, Nucleic Acids Res., 2019, 47, 1544–1556 CrossRef CAS.
- D. J. Y. Tan, P. Das, F. R. Winnerdy, K. W. Lim and A. T. Phan, Chem. Commun., 2020, 56, 5897–5900 RSC.
- T. Q. N. Nguyen, K. W. Lim and A. T. Phan, Sci. Rep., 2017, 7, 11969 CrossRef.
- S. Asamitsu, S. Obata, A. T. Phan, K. Hashiya, T. Bando and H. Sugiyama, Chem. – Eur. J., 2018, 24, 4428–4435 CrossRef CAS.
- Y. M. Vianney, P. Preckwinkel, S. Mohr and K. Weisz, Chem. – Eur. J., 2020, 26, 16910–16922 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |