Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Efficient and selective antibody modification with functionalised divinyltriazines†
Andrew J.
Counsell
 ,
Stephen J.
Walsh
,
Stephen J.
Walsh
 ,
Naomi S.
Robertson
,
Naomi S.
Robertson
 ,
Hannah F.
Sore
,
Hannah F.
Sore
 and
David R.
Spring
and
David R.
Spring
 *
*
Department of Chemistry, University of Cambridge, Cambridge, CB2 1EW, UK. E-mail: spring@ch.cam.ac.uk
First published on 10th June 2020
Abstract
A highly efficient disulfide rebridging strategy for the modification of monoclonal antibodies with substituted divinyltriazine linkers is reported. The reaction proceeds efficiently under mild conditions with near stoichiometric quantities of linker. This method of conjugation yields serum stable antibody conjugates with a controlled payload loading of 4.
Antibody–drug conjugates (ADCs) are a rapidly emerging class of therapeutics for the treatment of cancer.1,2 ADCs deliver potent cytotoxic agents to specific cells by exploiting the ability of the conjugated antibody to selectively target overexpressed cell-surface receptors. The success of the platform is attested by the upwards of 80 ADCs currently in clinical trials,1,3 and the eight ADCs that have been granted marketing approval by the Food and Drug Administration—the most recent being sacituzumab govitecan (Trodelvy®, Immunomedics) in April 2020 for the treatment of metastatic triple-negative breast cancer. Notwithstanding, many of the commonly adopted approaches to antibody conjugation have significant limitations. Common methods of attachment involve nucleophilic conjugation through multiple lysine or cysteine residues (generated by reduction of interchain disulfide bonds). Such stochastic modification strategies predominantly produce heterogeneous mixtures of conjugated antibody products, which accordingly suffer from unpredictable and inconsistent pharmacological profiles.4–6 Other approaches to attaining homogeneous constructs have been developed, such as the incorporation of engineered cysteine residues, unnatural amino acids, enzymatic modification, chemical linchpins, and affinity peptides—however may involve additional complexity.7–10
Site-selective disulfide rebridging has emerged as an alternative strategy towards the production of homogeneous ADCs.11 This method involves reduction of the interchain disulfides, followed by treatment with a linker which crosslinks the reactive thiols from the reduced disulfides, thus reforming the covalent bridge between the protein chains. Significant progress has been made in recent years in the development of disulfide rebridging linkers, with the toolbox of available reagents now including bissulfones,12 next generation maleimides,13–16 and pyridazinediones.17–19 Although these platforms constitute significant advances, new methodologies are required to further stimulate the development of homogeneous ADCs comprising native antibodies.
A notable issue with disulfide rebridging strategies is the potential for the formation of non-native cysteine bridges, resulting in an undesirable mixture of conjugates.20 Moreover, many modification strategies typically require a large excess of conjugation reagent, which may affect development and production costs depending upon the strategy adopted for toxin incorporation. Accordingly, the development of protocols with improved stoichiometry is a necessity.21–23 In light of these challenges, we sought to develop a disulfide rebridging platform that proceeds with increased efficiency and economy. We have recently demonstrated that divinylpyrimidines (DVPs) can be used for the efficient rebridging of disulfide bonds in antibodies and antibody fragments, producing stable conjugates with consistent drug-to-antibody ratio (DAR).24,25 The ability of divinyltriazines (DVTs) to function as cysteine stapling reagents in the development of functionalised macrocyclic peptide therapeutics has also been showcased.26 It was hypothesised that these more electron deficient DVT reagents may enable an efficient and economical synthesis of ADCs that forms a product with increased homogeneity. As such, we elected to adopt triazine 1 as a simple and easily accessible scaffold upon which to centre our antibody conjugation investigations (Fig. 1).
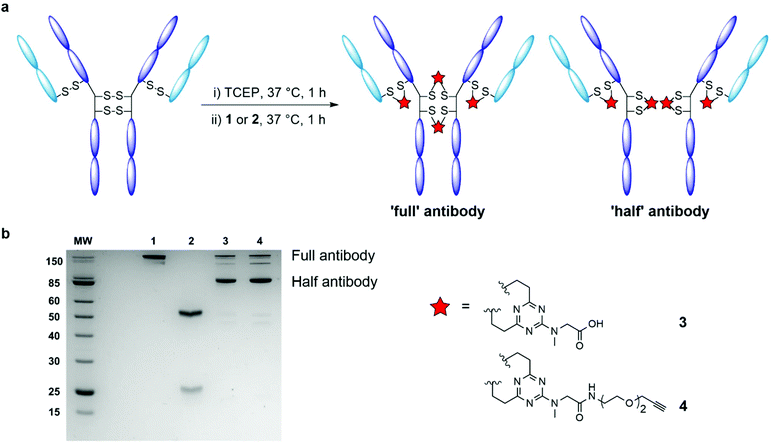
 | ||
| Fig. 1 Schematic representation of antibody disulfide rebridging with divinyltriazine linkers 1 and 2 described in this work. | ||
We began by examining the ability of model triazine 1 to rebridge the four interchain disulfide bonds found in an IgG1 antibody. Trastuzumab (Herceptin®) was selected for appraisal of the rebridging reaction. Commonly used in the evaluation of antibody bioconjugation strategies, trastuzumab is a monoclonal IgG1 antibody used in the treatment of HER2+ cancers, and is the antibody used in two approved ADCs (Kadcyla® and Enhertu®) and multiple ADCs currently undergoing clinical evaluation.27 The four solvent accessible interchain disulfide bonds of trastuzumab were reduced in Tris buffer (25 mM Tris HCl, 25 mM NaCl, 0.5 mM EDTA; pH 8.0) with an excess of the reducing agent tris(2-carboxyethyl) phosphine hydrochloride (TCEP; 10 eq.), to reveal eight free thiols. Complete reduction was confirmed by LC-MS and SDS-PAGE analysis (ESI Fig. S1†). The reduced antibody was then treated with DVT 1 (see ESI† for full synthetic details). Pleasingly, highly efficient rebridging of the disulfide bonds was observed with an excess of rebridging reagent (20 equivalents per IgG) over 4 hours at 37 °C. The reaction was monitored by LC-MS and SDS-PAGE analysis, which evidenced the formation of the correctly rebridged ‘full’ antibody, alongside the ‘half-antibody’ species formed through intrachain bridging of the heavy chain hinge region cysteines (Fig. 2a). A recent study has suggested that although so called ‘scrambling’ of antibody disulfide bonds has a slight impact on the Fc profile, the critical biophysical properties (e.g. antigen binding, aggregation) of the conjugate are largely unaffected.20 It is postulated that the scrambled conjugate remains effective as there is no change to the site of modification, the net DAR is unaffected, and the stability of the covalent linkage remains. Undesired ‘half-antibody’ formation has remained a persistent issue in the development of disulfide rebridging linkers, and so an optimisation study with triazine 1 was undertaken to determine conditions which favoured greater formation of the ‘full’ rebridged antibody conjugate. Sequential reduction of the antibody and linker addition across a range of conditions was undertaken, exploring the effect of varying the number of equivalents of linker, temperature, and duration of reaction (ESI Table S1†). Reduced trastuzumab was incubated with 10, 20, 40, or 80 equivalents of triazine 1, each at temperatures of 4, 20 and 37 °C. Each reaction was monitored over 8 hours. A number of observations were evident from this investigation. First, a significant dependence between temperature and the nature of bridging between the formed conjugates was found. Rebridging was found to occur with increased efficiency at higher temperatures (ESI Fig. S2†). Pleasingly, treatment of the reduced antibody with just 10 molar equivalents of DVT 1 for 1 hour at 37 °C was sufficient to afford excellent conversion to rebridged antibody. Extending the reaction time beyond 1 hour, or treatment with triazine 1 in excess of 10 equivalents did not offer appreciable improvements in rebridging efficiency. It was also found that removing excess TCEP and its by-products prior to addition of the DVT reagent yielded a marginal improvement in the conversion efficiency of the reaction from unreacted light and heavy chain to the rebridged antibody (ESI Fig. S2†). This suggested that TCEP or by-products produced as a result of it may slightly impede the rebridging reaction.
Further investigation of the excess of triazine 1 required was then undertaken. Conjugations to trastuzumab were conducted with between 4–10 equivalents of triazine 1. Although minor increases in light and heavy chain presence was observed via SDS-PAGE with lower concentrations of DVT, the rate of rebridging was still remarkable given the stoichiometry (ESI Fig. S3†). These results demonstrate that stoichiometric equivalents of triazine 1 can be used without significant detriment to the progression of the conjugation reaction.
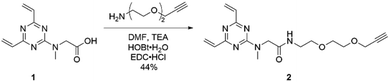
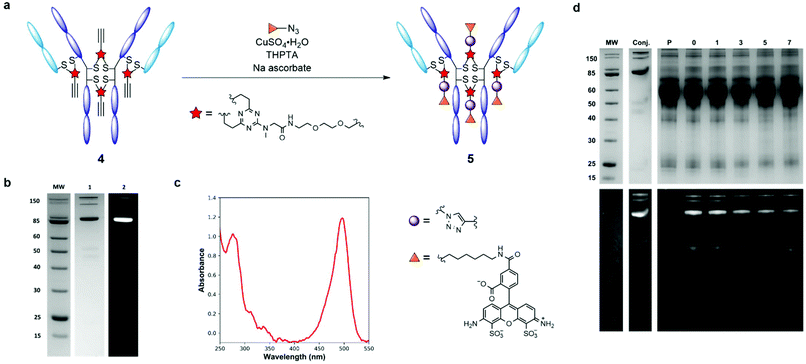
Having evaluated the ability of the divinyltriazine scaffold to rebridge the antibody interchain disulfides, slight modification of the linker was deemed necessary in order to introduce modular utility and a means of divergent functionalisation to the triazine following conjugation. A terminal alkyne was identified as an expedient choice, given the established nature of the copper-catalysed azide–alkyne cycloaddition (CuAAC) as one compatible with biologically relevant conditions.28–30 Accordingly, DVT 1 was reacted with 2-[2-(2-propynyloxy)ethoxy]ethylamine in the presence of N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC·HCl), 1-hydroxybenzotriazole hydrate (HOBt·H2O) and triethylamine (TEA) in DMF to yield alkynyl DVT 2 (Scheme 1). Subsequent treatment of reduced trastuzumab with triazine 2 (10 eq.) in Tris buffer for 1 h at 37 °C afforded the desired alkynyl antibody conjugate 4, as evidenced by LC-MS and SDS-PAGE analysis (Fig. 2 and ESI Fig. S4†). This conjugate was then reacted with AlexaFluor™ 488 azide in the presence of CuSO4·5H2O, tris(3-hydroxpyropyltriazolylmethyl)amine (THPTA), and sodium ascorbate (Fig. 3a). Excellent formation of the trastuzumab-AlexaFluor™ 488 conjugate was observed by SDS-PAGE, LC-MS and UV-Vis spectroscopy, with a measured fluorophore-antibody ratio (FAR) of 4.0 (Fig. 3b, c and ESI Fig. S5†).
Synthesis of antibody conjugate 5 allowed for the assessment of the plasma stability of DVT mediated bioconjugates. The fluorescent antibody conjugate 5 was incubated in reconstituted human plasma at 37 °C for one week. Aliquots were taken throughout the incubation period and were analysed via SDS-PAGE; in-gel fluorescence and Coomassie staining revealed no significant transfer of the AlexaFluor™ 488 label to plasma proteins, or loss of antibody chain fragments (Fig. 3d). This result demonstrates that DVT linkers can be employed to generate stable bioconjugates.
In conclusion, we have described the development of an efficient and economical strategy for the rebridging of antibody interchain disulfides. The linker equivalents required for a rapid production of plasma-stable antibody conjugates can be reduced to a near stoichiometric ratio, without significant detriment to reaction efficiency. The platform incorporates a vector that facilitates bioorthogonal divergent functionalisation (CuAAC). As such, the DVT scaffold provides a useful addition to the toolbox of reagents available for selective disulfide rebridging of proteins.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to AstraZeneca for providing trastuzumab for this research. A. J. C acknowledges support from Trinity College, Cambridge. D. R. S acknowledges support from the Engineering and Physical Sciences Research Council (EP/P020291/1) and Royal Society (Wolfson Research Merit Award). The Spring lab acknowledges general lab support from the EPSRC, BBSRC, MRC and Royal Society.Notes and references
- A. Beck, L. Goetsch, C. Dumontet and N. Corvaïa, Nat. Rev. Drug Discovery, 2017, 16, 315–337 CrossRef CAS PubMed.
- P. J. Carter and G. A. Lazar, Nat. Rev. Drug Discovery, 2018, 17, 197–223 CrossRef CAS PubMed.
- M. Abdollahpour-Alitappeh, M. Lotfinia, T. Gharibi, J. Mardaneh, B. Farhadihosseinabadi, P. Larki, B. Faghfourian, K. S. Sepehr, K. Abbaszadeh-Goudarzi, G. Abbaszadeh-Goudarzi, B. Johari, M. R. Zali and N. Bagheri, J. Cell. Physiol., 2019, 234, 5628–5642 CrossRef CAS PubMed.
- J. R. Junutula, H. Raab, S. Clark, S. Bhakta, D. D. Leipold, S. Weir, Y. Chen, M. Simpson, S. P. Tsai, M. S. Dennis, Y. Lu, Y. G. Meng, C. Ng, J. Yang, C. C. Lee, E. Duenas, J. Gorrell, V. Katta, A. Kim, K. McDorman, K. Flagella, R. Venook, S. Ross, S. D. Spencer, W. Lee Wong, H. B. Lowman, R. Vandlen, M. X. Sliwkowski, R. H. Scheller, P. Polakis and W. Mallet, Nat. Biotechnol., 2008, 26, 925–932 CrossRef CAS PubMed.
- B. Q. Shen, K. Xu, L. Liu, H. Raab, S. Bhakta, M. Kenrick, K. L. Parsons-Reponte, J. Tien, S. F. Yu, E. Mai, D. Li, J. Tibbitts, J. Baudys, O. M. Saad, S. J. Scales, P. J. McDonald, P. E. Hass, C. Eigenbrot, T. Nguyen, W. A. Solis, R. N. Fuji, K. M. Flagella, D. Patel, S. D. Spencer, L. A. Khawli, A. Ebens, W. L. Wong, R. Vandlen, S. Kaur, M. X. Sliwkowski, R. H. Scheller, P. Polakis and J. R. Junutula, Nat. Biotechnol., 2012, 30, 184–189 CrossRef CAS PubMed.
- K. J. Hamblett, P. D. Senter, D. F. Chace, M. M. C. Sun, J. Lenox, C. G. Cerveny, K. M. Kissler, S. X. Bernhardt, A. K. Kopcha, R. F. Zabinski, D. L. Meyer and J. A. Francisco, Clin. Cancer Res., 2004, 10, 7063–7070 CrossRef CAS PubMed.
- K. Tsuchikama and Z. An, Protein Cell, 2018, 9, 33–46 CrossRef CAS PubMed.
- V. Chudasama, A. Maruani and S. Caddick, Nat. Chem., 2016, 8, 114–119 CrossRef CAS PubMed.
- K. Yamada, N. Shikida, K. Shimbo, Y. Ito, Z. Khedri, Y. Matsuda and B. A. Mendelsohn, Angew. Chem., Int. Ed., 2019, 58, 5592–5597 CrossRef CAS PubMed.
- S. R. Adusumalli, D. G. Rawale, U. Singh, P. Tripathi, R. Paul, N. Kalra, R. K. Mishra, S. Shukla and V. Rai, J. Am. Chem. Soc., 2018, 140, 15114–15123 CrossRef CAS PubMed.
- N. Forte, V. Chudasama and J. R. Baker, Drug Discovery Today: Technol., 2018, 30, 11–20 CrossRef PubMed.
- G. Badescu, P. Bryant, M. Bird, K. Henseleit, J. Swierkosz, V. Parekh, R. Tommasi, E. Pawlisz, K. Jurlewicz, M. Farys, N. Camper, X. Sheng, M. Fisher, R. Grygorash, A. Kyle, A. Abhilash, M. Frigerio, J. Edwards and A. Godwin, Bioconjugate Chem., 2014, 25, 1124–1136 CrossRef CAS PubMed.
- M. E. B. Smith, F. F. Schumacher, C. P. Ryan, L. M. Tedaldi, D. Papaioannou, G. Waksman, S. Caddick and J. R. Baker, J. Am. Chem. Soc., 2010, 132, 1960–1965 CrossRef CAS PubMed.
- J. P. M. Nunes, M. Morais, V. Vassileva, E. Robinson, V. S. Rajkumar, M. E. B. Smith, R. B. Pedley, S. Caddick, J. R. Baker and V. Chudasama, Chem. Commun., 2015, 51, 10624–10627 RSC.
- F. F. Schumacher, J. P. M. Nunes, A. Maruani, V. Chudasama, M. E. B. Smith, K. A. Chester, J. R. Baker and S. Caddick, Org. Biomol. Chem., 2014, 12, 7261–7269 RSC.
- F. Bryden, C. Martin, S. Letast, E. Lles, I. Viéitez-Villemin, A. Rousseau, C. Colas, M. Brachet-Botineau, E. Allard-Vannier, C. Larbouret, M. C. Viaud-Massuard and N. Joubert, Org. Biomol. Chem., 2018, 16, 1882–1889 RSC.
- A. Maruani, M. E. B. Smith, E. Miranda, K. A. Chester, V. Chudasama and S. Caddick, Nat. Commun., 2015, 6, 6645 CrossRef CAS PubMed.
- M. T. W. Lee, A. Maruani, D. A. Richards, J. R. Baker, S. Caddick and V. Chudasama, Chem. Sci., 2017, 8, 2056–2060 RSC.
- M. T. W. Lee, A. Maruani, J. R. Baker, S. Caddick and V. Chudasama, Chem. Sci., 2016, 7, 799–802 RSC.
- C. Bahou, E. A. Love, S. Leonard, R. J. Spears, A. Maruani, K. Armour, J. R. Baker and V. Chudasama, Bioconjugate Chem., 2019, 30, 1048–1054 CrossRef CAS PubMed.
- R. M. Devay, K. Delaria, G. Zhu, C. Holz, D. Foletti, J. Sutton, G. Bolton, R. Dushin, C. Bee, J. Pons, A. Rajpal, H. Liang, D. Shelton, S. H. Liu and P. Strop, Bioconjugate Chem., 2017, 28, 1102–1114 CrossRef CAS PubMed.
- B. Stump and J. Steinmann, in Methods in Molecular Biology, ed. L. Ducry, Humana Press Inc., 2013, vol. 1045, pp. 235–248 Search PubMed.
- Credence Research, Global Antibody Drug Conjugate Market to Reach Worth USD 29.3 Bn by 2024: Promising Drug Pipeline to Drive the Market, 2017 Search PubMed.
- S. J. Walsh, S. Omarjee, W. R. J. D. Galloway, T. T.-L. Kwan, H. F. Sore, J. S. Parker, M. Hyvönen, J. S. Carroll and D. R. Spring, Chem. Sci., 2019, 10, 694–700 RSC.
- S. J. Walsh, J. Iegre, H. Seki, J. D. Bargh, H. F. Sore, J. S. Parker, J. S. Carroll and D. R. Spring, Org. Biomol. Chem., 2020, 18, 4224–4230 RSC.
- N. S. Robertson, S. J. Walsh, E. Fowler, M. Yoshida, S. M. Rowe, Y. Wu, H. F. Sore, J. S. Parker and D. R. Spring, Chem. Commun., 2019, 55, 9499–9502 RSC.
- N. Pondé, P. Aftimos and M. Piccart, Curr. Treat. Options Oncol., 2019, 20, 1–22 CrossRef PubMed.
- M. Z. C. Hatit, L. F. Reichenbach, J. M. Tobin, F. Vilela, G. A. Burley and A. J. B. Watson, Nat. Commun., 2018, 9, 1–7 CrossRef CAS PubMed.
- C. Besanceney-Webler, H. Jiang, T. Zheng, L. Feng, D. Soriano del Amo, W. Wang, L. M. Klivansky, F. L. Marlow, Y. Liu and P. Wu, Angew. Chem., Int. Ed., 2011, 50, 8051–8056 CrossRef CAS PubMed.
- V. Hong, S. I. Presolski, C. Ma and M. G. Finn, Angew. Chem., Int. Ed., 2009, 48, 9879–9883 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ob01002b |
| This journal is © The Royal Society of Chemistry 2020 |