 Open Access Article
Open Access ArticlePhysical magnification of objects
Edward S.
Boyden

Media Lab, McGovern Institute, Koch Institute, Department of Biological Engineering, and Department of Brain and Cognitive Sciences, MIT, Cambridge, MA 02139, USA. E-mail: esb@media.mit.edu
First published on 2nd October 2018
Abstract
For hundreds of years, scientists have aimed photons, electrons, and X-rays at objects in order to obtain magnified images of them, revealing their micro- and nanoscale architecture, and revolutionizing every branch of the sciences. Recently a new modality of magnification was discovered – physical magnification of objects. For objects with certain properties, such as biological specimens, one can synthesize throughout them a dense, even network of swellable polymer, so that further chemical processing steps will isotropically expand the polymer, and thus the object, manyfold in physical dimension. We here explore the principles governing this new modality of magnification, such as the ability to use inexpensive, ubiquitous imaging hardware to perform high-resolution imaging of objects, and the ability to decrowd molecules within an object for detailed in situ chemical analysis.
Light microscopes obtain images of objects by delivering light to a specimen, and using lenses to magnify the resulting optical images of the specimen. The resolution of the microscope is limited by the wavelength of the light, meaning that features smaller than a few hundred nanometers are not resolvable. Super-resolution methods were developed that can break this so-called diffraction limit, but they are slow, expensive, and/or complex to use. Recently a new modality of magnification was discovered, namely the direct physical magnification of objects, which enables nanoscale features to be resolved with ordinary, ubiquitous light microscopes (Fig. 1).1 For physical objects that are porous, such as biological specimens, it is possible to synthesize (Fig. 1A) throughout such objects a dense, crosslinked, evenly synthesized network of swellable polymer – for example, a hydrogel of a polyelectrolyte such as sodium polyacrylate – covalently anchoring key molecules of interest within the object to the polymer network. The spacing between the polymer threads (or mesh size) should ideally be comparable to the spacing between the molecules of interest, or at least the resolution desired. Then, one can mechanically disrupt the endogenous interactions between the molecules of interest, or even destroy molecules that are not of interest, e.g. through heat, enzymatic, or chemical treatment. Finally, adding water causes the hydrogel to swell, pulling the molecules of interest apart from one another via the anchors. After expanding an object by ∼4 fold in linear dimension (Fig. 1B and C), a light microscope with diffraction limit 300 nm would have an effective resolution of ∼300/4 ∼ 70 nm – enabling nanoscopy on ordinary microscopes commonly available in science. This strategy is called expansion microscopy (ExM).1 Given the nanoscale spacing between polymer threads in such samples, the fundamental limit of such expansion processes might be as small as 5–10 nm.8 Given that the final object is ∼99% water, having been swollen in water, it is effectively completely transparent by the end of the process (Fig. 1D), which further facilitates the power of fast, diffraction-limited methods of imaging such as light-sheet microscopy.7
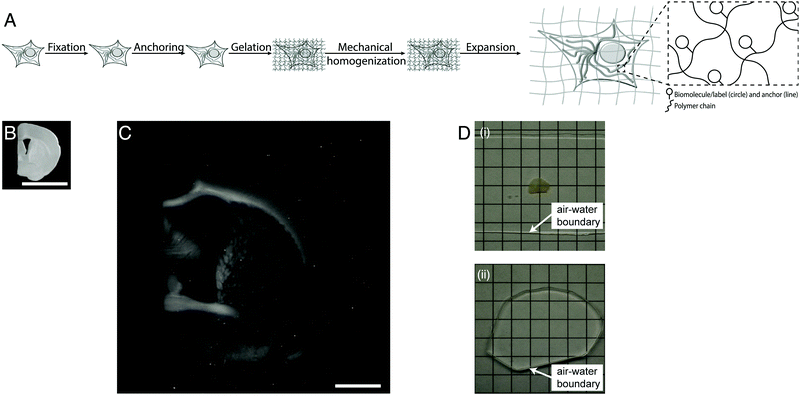 | ||
| Fig. 1 Physical magnification of objects: workflow and example. (A) The workflow of expansion microscopy, as applied to biological specimens. The biological specimen is first preserved (“fixed”) through a chemical process, then key biomolecules or labels within the specimen are covalently equipped with anchor molecules (“anchoring”). A dense network of swellable polyelectrolyte hydrogel is then synthesized throughout the specimen, binding to the anchor molecules, and with polymer spacing comparable to the spacing between biomolecules or labels of interest (“gelation”). Finally, the specimen is chemically treated to soften the mechanical properties (“mechanical homogenization”) and then finally expanded by adding an appropriate solvent such as water (“expansion”). (B) Specimen of preserved mouse brain. (C) The same specimen as in B, but after expansion as in A. (D) Expansion of a specimen of brain (i) makes the specimen transparent (ii), as in its final state it is mostly water. Scale bars: (B) and (C) 5 mm, in physical size units. Adapted from ref. 21. | ||
ExM owes a debt to two lines of research that go back many decades. In the early 1980s, the phase transition physics underlying changes in volume of swellable polyelectrolyte hydrogels was worked out,2 and around the same time, it was discovered by biologists that polymer hydrogel embedding of tissues (by soaking specimens in a solution of acrylamide monomer, which in turn would undergo free radical polymerization, resulting in a polyacrylamide hydrogel permeating throughout a preserved specimen) could be used to facilitate biological imaging.3 To date this strategy has been primarily applied to preserved biological cells and tissues, which are highly porous, with proteins4–6 or nucleic acids7 being anchored to the polyelectrolyte hydrogel via commercially available anchoring molecules. One can expand an object over and over, forming a second polymer in the space opened up by a first round of expansion, for added resolution (∼4 × 4 = 16× magnification would enable ∼300/16 ∼ 20 nm resolution).8 Recently expansion has been applied to other kinds of specimens, such as crystals of proteins, which were expanded and contracted, and assessed by X-ray imaging.9 Recently an unrelated chemistry platform, relying on DNA origami, was used to expand multimeric protein complexes so that individual protein building blocks could be optically resolved, for structural analysis.10 This highlights that the concept of expansion is not limited only to the use of polymer hydrogels; the initial discovery that it was possible to isotropically expand a biological specimen has kicked off a lot of excitement about alternative ways of doing this. For example, a non-polymeric strategy using just aqueous chemical solutions was recently shown to be capable of expanding entire mouse brains by ∼2×.11 Nevertheless the clear rationale, high magnification, and extensive empirical validation (e.g., by comparing to older technologies or to known “ground truth” structures,1,5,6,8,12,13 it has been shown that the distortion is low, perhaps a few percent over length scales of tens to hundreds of microns) behind polymer hydrogel expansion microscopy has made it the most popular to date. So far this strategy has not been applied to completely non-biological objects, but in principle it could be applied to any porous object that is amenable to the steps described above. In biology, however, it has rapidly seen adoption, with simple protocols of use emerging,14 and with papers rapidly appearing that apply expansion microscopy to human cancer specimens,13 the brain of the fruit fly,15 the larval zebrafish,12 human brain tissue,16 planaria,17 the pathogen Giardia lamblia,18 and many other kinds of specimen. Expansion of human pathology specimens has even been shown to boost the performance of machine learning algorithms in early cancer detection,13 highlighting the potential of expansion imaging to make subtle signatures of disease into highly visible features. The simplicity of the protocol, and the fact that it enables nanoscale imaging on hardware that scientists already have, have contributed to the rapid spread of the technology.
Expansion of an object can be used to improve the resolution of any microscope, in principle. For example, the application of expansion to bacteria was used to make them detectable by inexpensive, modified webcams,19 which might make diagnosis of disease or detection of pathogens very cheap. The expansion of brain circuitry makes neural wiring easier to trace.5 A perhaps nonobvious advantage of expansion is that it decrowds molecules within a specimen from one another, making room for interesting chemical reactions such as amplification, hybridization, sequencing, and other analytical chemistries. Already it has been shown that expansion of a cell, by decrowding messenger RNA (mRNA) molecules from each other, could facilitate their more accurate counting, and furthermore made room for amplification of the fluorescence through the fluorescence-amplifying hybridization chain reaction (HCR).7 This decrowding also greatly facilitates coded serial hybridization methods like MERFISH, which can read out an exponential amount of information in a linear number of steps, by applying carefully designed hybridization probes in multiple rounds.20 The ability to decrowd proteins from each other, if achievable in a controlled way, may enable epitopes that cannot be detected by antibody binding, to be tagged because the decrowding makes room around each protein for antibody approach. In summary, the recent discovery that specimens such as biological samples can be physically magnified by a material science process is opening up frontiers in the nanoscale mapping of extended objects, in the inexpensive probing of molecular complexes and other nanoscale features, and in the multiplexed analysis of biomolecules through decrowding-enhanced in situ chemistry. In the future, greater degrees of physical magnification and higher isotropy through new polymeric chemistries, as well as novel and powerful kinds of in situ analysis chemistry, may make the expansion microscopy toolset an even more powerful toolbox for understanding the configuration of molecules throughout cells and cells throughout tissues, and perhaps reveal the molecular configuration of other kinds of specimen as well.
Conflicts of interest
E. S. B. is an inventor on multiple patents related to expansion microscopy, and also has co-founded a company to pursue clinical and translational applications of expansion microscopy.References
- F. Chen, P. W. Tillberg and E. S. Boyden, Expansion Microscopy, Science, 2015, 347, 543–548 CrossRef CAS PubMed.
- T. Tanaka, D. Fillmore, S. T. Sun, I. Nishio, G. Swislow and A. Shah, Phase Transitions in Ionic Gels, Phys. Rev. Lett., 1980, 45, 1636–1639 CrossRef CAS.
- P. Hausen and C. Dreyer, The Use of Polyacrylamide as an Embedding Medium for Immunohistochemical Studies of Embryonic Tissues, Stain Technol., 1981, 56, 287–293 CrossRef CAS PubMed.
- T. Ku, J. Swaney, J.-Y. Park, A. Albanese, E. Murray, J. H. Cho, Y.-G. Park, V. Mangena, J. Chen and K. Chung, Multiplexed and Scalable Super-Resolution Imaging of Three-Dimensional Protein Localization in Size-Adjustable Tissues, Nat. Biotechnol., 2016, 34, 973–981 CrossRef CAS PubMed.
- P. W. Tillberg, F. Chen, K. D. Piatkevich, Y. Zhao, C.-C. Yu (Jay), B. P. English, L. Gao, A. Martorell, H.-J. Suk, F. Yoshida, M. Ellen, D. H. R. DeGennaro, G. Gong, U. Seneviratne, S. R. Tannenbaum, R. Desimone, D. Cai and E. S. Boyden, Protein-Retention Expansion Microscopy of Cells and Tissues Labeled Using Standard Fluorescent Proteins and Antibodies, Nat. Biotechnol., 2016, 34, 987–992 CrossRef CAS PubMed.
- T. J. Chozinski, A. R. Halpern, H. Okawa, H.-J. Kim, G. J. Tremel, R. O. L. Wong and J. C. Vaughan, Expansion Microscopy with Conventional Antibodies and Fluorescent Proteins, Nat. Methods, 2016, 13, 1–7 CrossRef PubMed.
- F. Chen, A. T. Wassie, A. J. Cote, A. Sinha, S. Alon, E. R. D. Shoh Asano, J.-B. Chang, A. Marblestone, G. M. Church, A. Raj and E. S. Boyden, Nanoscale Imaging of RNA with Expansion Microscopy, Nat. Methods, 2016, 13, 679–684 CrossRef CAS PubMed.
- J.-B. Chang, F. Chen, Y.-G. Yoon, E. E. Jung, H. Babcock, J. S. Kang, S. Asano, H.-J. Suk, N. Pak, P. W. Tillberg, A. T. Wassie, D. Cai and E. S. Boyden, Iterative Expansion Microscopy, Nat. Methods, 2017, 14, 593–599 CrossRef CAS PubMed.
- L. Zhang, J. B. Bailey, R. H. Subramanian and F. A. Tezcan, Hyperexpandable, Self-Healing Macromolecular Crystals with Integrated Polymer Networks, Nature, 2018, 557, 86–91 CrossRef CAS PubMed.
- J. Molle, L. Jakob, J. Bohlen, M. Raab and P. Tinnefeld, Dina Grohmann Towards Structural Biology with Super-Resolution Microscopy, Nanoscale, 2018, 10, 16416–16424 RSC.
- T. C. Murakami, T. Mano, S. Saikawa, S. A. Horiguchi, D. Shigeta, K. Baba, H. Sekiya, Y. Shimizu, K. F. Tanaka, H. Kiyonari, M. Iino, H. Mochizuki, K. Tainaka and H. R. Ueda, A Three-Dimensional Single-Cell-Resolution Whole-Brain Atlas Using CUBIC-X Expansion Microscopy and Tissue Clearing, Nat. Neurosci., 2018, 21, 625–637 CrossRef CAS PubMed.
- L. Freifeld, I. Odstrcil, D. Förster, A. Ramirez, J. A. Gagnon, O. Randlett, E. K. Costa, S. Asano, O. T. Celiker, R. Gao, D. A. Martin-Alarcon, P. Reginato, C. Dick, L. Chen, D. Schoppik, F. Engert and E. S. Boyden, Expansion Microscopy of Zebrafish for Neuroscience and Developmental Biology Studies, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E10799–E10808 CrossRef CAS PubMed.
- Y. Zhao, O. Bucur, H. Irshad, F. Chen, A. Weins, A. L. Stancu, E.-Y. Oh, M. DiStasio, V. Torous, B. Glass, I. E. Stillman, S. J. Schnitt, A. H. Beck and E. S. Boyden, Nanoscale Imaging of Clinical Specimens Using Pathology-Optimized Expansion Microscopy, Nat. Biotechnol., 2017, 35, 757–764 CrossRef CAS PubMed.
- ExpansionMicroscopy.Org: Physical Specimen Expansion Enabling 3-D Large Volume, Nanoscale Imaging http://expansionmicroscopy.org/.
- T. J. Mosca, D. J. Luginbuhl, I. E. Wang and L. Luo, Presynaptic LRP4 Promotes Synapse Number and Function of Excitatory CNS Neurons, eLife, 2017, 6, e27347 CrossRef PubMed.
- T. Deshpande, T. Li, M. K. Herde, A. Becker, H. Vatter, M. K. Schwarz, C. Henneberger, C. Steinhäuser and P. Bedner, Subcellular Reorganization and Altered Phosphorylation of the Astrocytic Gap Junction Protein Connexin43 in Human and Experimental Temporal Lobe Epilepsy, Glia, 2017, 65, 1809–1820 CrossRef PubMed.
- I. E. Wang, S. W. Lapan, M. L. Scimone, T. R. Clandinin and P. W. Reddien, Hedgehog Signaling Regulates Gene Expression in Planarian Glia, eLife, 2016, 5, e16996 CrossRef PubMed.
- A. R. Halpern, G. C. M. Alas, T. J. Chozinski, A. R. Paredez and J. C. Vaughan, Hybrid Structured Illumination Expansion Microscopy Reveals Microbial Cytoskeleton Organization, ACS Nano, 2017, 11, 12677–12686 CrossRef CAS PubMed.
- Y. S. Zhang, J.-B. Chang, M. M. Alvarez, G. Trujillo-de Santiago, J. Aleman, B. Batzaya, V. Krishnadoss, A. A. Ramanujam, M. Kazemzadeh-Narbat, F. Chen, P. W. Tillberg, M. R. Dokmeci, E. S. Boyden and A. Khademhosseini, Hybrid Microscopy: Enabling Inexpensive High-Performance Imaging through Combined Physical and Optical Magnifications, Sci. Rep., 2016, 6, 22691 CrossRef CAS PubMed.
- G. Wang, J. R. Moffitt and X. Zhuang, Multiplexed Imaging of High-Density Libraries of RNAs with MERFISH and Expansion Microscopy, Sci. Rep., 2018, 8, 4847 CrossRef PubMed.
- S. Alon, G. Huynh and E. S. Boyden, Expansion Microscopy: Enabling Single Cell Analysis in Intact Biological Systems, FEBS J., 2018 DOI:10.1111/febs.14597.
| This journal is © The Royal Society of Chemistry 2019 |
