 Open Access Article
Open Access ArticleNanoscale chemical analysis of 2D molecular materials using tip-enhanced Raman spectroscopy
Dušan
Mrđenović
 ,
Zhen-Feng
Cai
,
Zhen-Feng
Cai
 ,
Yashashwa
Pandey
,
Yashashwa
Pandey
 ,
Giovanni Luca
Bartolomeo
,
Renato
Zenobi
,
Giovanni Luca
Bartolomeo
,
Renato
Zenobi
 * and
Naresh
Kumar
* and
Naresh
Kumar
 *
*
Department of Chemistry and Applied Biosciences, ETH Zurich, CH-8093 Zurich, Switzerland. E-mail: zenobi@org.chem.ethz.ch; kumar@org.chem.ethz.ch
First published on 21st December 2022
Abstract
Two-dimensional (2D) molecular materials have attracted immense attention due to their unique properties, promising a wide range of exciting applications. To understand the structure–property relationship of these low-dimensional materials, sensitive analytical tools capable of providing structural and chemical characterisation at the nanoscale are required. However, most conventional analytical techniques fail to meet this challenge, especially in a label-free and non-destructive manner under ambient conditions. In the last two decades, tip-enhanced Raman spectroscopy (TERS) has emerged as a powerful analytical technique for nanoscale chemical characterisation by combining the high spatial resolution of scanning probe microscopy and the chemical sensitivity and specificity of surface-enhanced Raman spectroscopy. In this review article, we provide an overview of the application of TERS for nanoscale chemical analysis of 2D molecular materials, including 2D polymers, biomimetic lipid membranes, biological cell membranes, and 2D reactive systems. The progress in the structural and chemical characterisation of these 2D materials is demonstrated with key examples from our as well as other laboratories. We highlight the unique information that TERS can provide as well as point out the common pitfalls in experimental work and data interpretation and the possible ways of averting them.
1. Introduction
Two-dimensional (2D) molecular materials are composed of a single or few layers of molecules held together via covalent bonds and/or inter- and intra-layer van der Waals interactions. Such a structural arrangement gives them unique properties compared to their bulk counterparts. For example, whilst synthetic polymers have been around for more than a century, 2D polymers (2DPs) were synthesised for the first time in 2005.1 2DPs are covalently linked molecular networks consisting of periodic bonding structures, whose physiochemical characteristics are determined by the chemistry of monomeric building blocks.2 Compared to bulk polymers, 2DPs offer greater flexibility in tuning physiochemical properties including composition, porosity, and surface functionalisation,3–5 making them promising candidates for potential applications in organic electronics, heterogeneous catalysis, and nanosieves.6,7Similarly, owing to their nature-inspired design, 2D biomimetic molecular systems such as supported lipid membranes have proven to be immensely useful in a wide variety of areas including cancer treatment, drug delivery, tissue engineering, sensor design, etc.8 Supported monolayer and bilayer lipid membranes are an interesting class of biomimetic systems in which molecules are held together by intra- and inter-layer van der Waals interactions. These systems are not only useful for practical applications such as biosensing,9 but also have an important role in fundamental research as cell membrane models. These model systems have less complex molecular composition than real cell membranes, which allows for studying cellular processes more efficiently.
To obtain a clear understanding of the structure–property relationship in 2D molecular materials, structural and chemical characterisation at the nanometre length scale are required. However, this is a challenging task for most conventional analytical techniques because of at least one of the following reasons: (i) lack of chemical information, (ii) low sensitivity, (iii) lack of nanoscale spatial resolution, and (iv) sample damage. For example, atomic force microscopy (AFM)10 and scanning tunnelling microscopy (STM)11 can provide a very high spatial resolution reaching the sub-nm scale, but have no or very limited chemical characterisation ability. Conversely, infrared (IR)12 and Raman13 spectroscopy can provide chemical information with high molecular specificity, but their diffraction-limited spatial resolution prevents them from revealing nanoscale properties.
In the last few decades, nanoscale characterisation techniques have emerged as a solution to the low spatial resolution and sensitivity of the conventional analytical tools. For example, nanoscale secondary ion mass spectrometry can perform chemical analysis with a spatial resolution of up to 50 nm.14 However, it cannot operate under ambient conditions, is destructive, and can only be used to detect elements or very small molecular fragments (such as CH, CN, etc.) since the high spatial resolution demands the use of hard ionisation. On the other hand, super-resolution fluorescence microscopy can also provide a spatial resolution of up to 10–20 nm.15 However, fluorophore labelling is required, which can perturb molecular behaviour and only the labelled molecules can be observed whilst the rest of the sample remains undetectable. AFM–IR is a more recent nanoscale chemical characterisation tool that does not require labelling, is non-destructive, and can be performed under ambient conditions. However, the resolution is typically limited to a few tens of nm and the experiments in an aqueous environment are infeasible due to the strong IR light absorption of water.16
In the last two decades, tip-enhanced Raman spectroscopy (TERS) has emerged as a powerful analytical technique that combines the high spatial resolution of AFM or STM and the chemical sensitivity and specificity of surface-enhanced Raman spectroscopy (SERS) to study surface chemistry at the nanoscale in a label-free and non-destructive manner.16–19 TERS has been successfully applied for nanoscale analysis in several areas of scientific research such as 2D materials including graphene, MoS2 and WSe2 and graphitic sheets,20–24 on-surface decomposition chemistry,25 polymer blends,26 photocatalytic reactions,27–29 organic solar cells,30 supported lipid membranes,31 biomembranes,32 and biological cells.33 In TERS, Raman scattering from a nanoscopic volume of the sample is plasmonically enhanced by several orders of magnitude owing to a highly intense and localised electromagnetic (EM) field, which is generated at the apex of a metallic SPM probe via a combination of localised surface plasmon resonance and lightening rod effect.34–36 There are three TERS configurations based on the direction of laser illumination at the sample surface, as schematically illustrated in Fig. 1. In the bottom-illumination setup (Fig. 1a), an objective with >1 numerical aperture (NA) focuses an excitation laser on the sample surface through a transparent substrate. In this configuration, a radially polarised laser beam is typically employed as it provides a tighter focus and a more intense longitudinal electric field along the probe axis, leading to a >4 times stronger EM enhancement compared to a linearly polarised beam.37 In the top illumination setup (Fig. 1b), the laser is focused from the top onto a tilted probe apex placed on the sample. In the side illumination setup (Fig. 1c), a long working distance objective lens focuses a linearly polarised beam onto the tip–sample interface from the side. In this case, even a relatively lower NA objective lens is able to provide a strong EM enhancement and localisation, when the laser polarisation is set parallel to the probe axis.
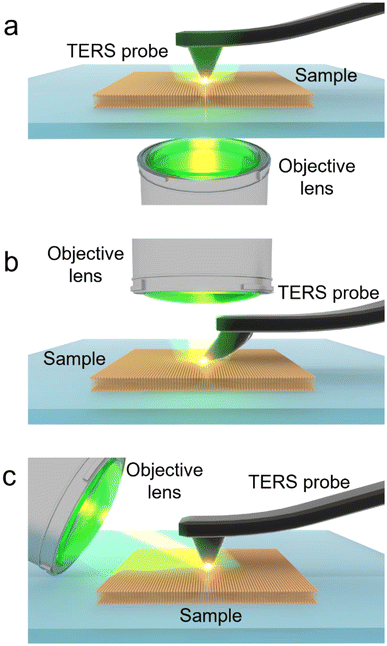 | ||
| Fig. 1 Schematic illustration of different TERS setups. (a) Bottom illumination, (b) top illumination and (c) side illumination. | ||
In this review article, we present an overview of the latest research on the nanoscale analysis of 2D molecular materials using TERS. We will primarily focus on four types of materials: (i) 2D polymers, (ii) biomimetic lipid membranes, (iii) biological membranes, and (iv) 2D reactive systems. Our aim is to demonstrate the unique information that TERS can provide at the nanoscale from these molecular 2D systems, pinpoint the typical pitfalls of experimental work and data interpretation as well as propose possible strategies for averting them.
2. 2D polymers
The first TERS study of 2DPs was conducted by Opilik et al. on a monomolecular sheet produced from amphiphilic monomers by anthracene photodimerization at an air–water interface, transferred onto a substrate via the Langmuir–Blodgett (LB) technique.38 TERS measurements indicated a successful dimerization and confirmed that the reaction mechanism is a [4 + 4] cycloaddition, rulling out the [4 + 2] cycloaddition. TERS investigation of a 0.8 nm-thick 2D covalent polyimine monolayer produced from triamine and dialdehyde by dynamic imine chemistry in a Langmuir trough and transferred on a substrate using Langmuir–Schaeffer (LS) technique was performed by Dai et al.39 Optical microscopy, electron microscopy, and AFM were used to confirm the presence of a monolayer, while the sample chemistry was studied using TERS and confocal Raman spectroscopy supported by density functional theory (DFT) calculations. TERS measurements indicated the presence of imine bonds and the absence of end groups, suggesting that the monomer successfully polymerised into an imine-linked monolayer. The authors also investigated nanoscale defects as their presence can significantly alter the physical, mechanical, optoelectronic, and chemical properties of the 2DP.40 For this, an acetylenic moiety was introduced into one monomer. TERS surface selection rules (the more aligned the molecule's polarizability tensor with the probe axis, the higher the TERS signal) allowed probing of the orientation of the acetylenic moiety. Since all other structural elements of the 2DP lie flat on the substrate surface, the acetylenic moiety could either lie flat (i.e. being incorporated into the polymer monolayer) or it could have a certain degree of freedom, thus indicating the presence of nanodefects, as schematically illustrated in Fig. 2a. Using TERS imaging (Fig. 2b), 98% of the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C bonds in the 2DP monolayer were estimated to be parallel to the substrate surface, i.e., they were locked inside the 2DP network. Moreover, STM and TERS analyses of the 2DP monolayer also showed that the monolayer did not rupture or become defective when deposited on Au(111) terrace edges (Fig. 2c–e). Instead, molecules present at terrace edges reoriented, giving rise to a higher TERS signal.
C bonds in the 2DP monolayer were estimated to be parallel to the substrate surface, i.e., they were locked inside the 2DP network. Moreover, STM and TERS analyses of the 2DP monolayer also showed that the monolayer did not rupture or become defective when deposited on Au(111) terrace edges (Fig. 2c–e). Instead, molecules present at terrace edges reoriented, giving rise to a higher TERS signal.
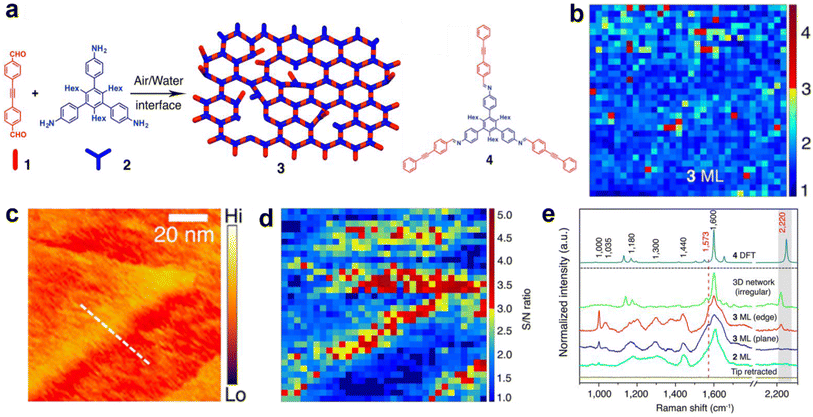 | ||
Fig. 2 (a) Schematic diagram illustrating the formation of a 2D covalent polyimine monolayer (3) from dialdehyde (1) and triamine (2) monomers. The structure of the imine compound (4) is also shown. (b) TERS image of the intact polyimine monolayer. (c) STM and (d) TERS images of the polyimine monolayer on the terrace of an Au(111) surface. All TERS images were constructed using the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C Raman signal at 2220 cm−1. (e) TERS spectra of the triamine monolayer (2 ML), polymer monolayer on the plane (3 ML plane) and edge (3 ML edge) of the Au(111) terrace, an irregular polyimine network (3D network irregular) and the DFT calculated spectra of a imine model compound (4 DFT). This figure has been adapted from ref. 40 with permission from the American Chemical Society, copyright 2018. C Raman signal at 2220 cm−1. (e) TERS spectra of the triamine monolayer (2 ML), polymer monolayer on the plane (3 ML plane) and edge (3 ML edge) of the Au(111) terrace, an irregular polyimine network (3D network irregular) and the DFT calculated spectra of a imine model compound (4 DFT). This figure has been adapted from ref. 40 with permission from the American Chemical Society, copyright 2018. | ||
Orientation of the fluorinated hydrocarbon (carboxylic-fantrip) molecules in 2DPs, which were either spin-coated or transferred by LS technique onto a Au(111) surface was studied using TERS by Shao et al.41 DFT calculations predicted a high ratio of the Raman vibrational modes at 1600 cm−1 and 1445 cm−1 (I1600/I1445) for molecules lying flat on the substrate and a low ratio in the upright position. Based on the I1600/I1445 ratio, TERS measurements of the molecules in the LS-transferred monolayer technique indicated a more rigid molecular orientation being stacked in a face-to-face configuration due to π–π interactions. Conversely, molecules in the spin-coated monolayers were found to adopt a random configuration. TERS images showed a uniform I1600/I1445 ratio across the LS-transferred samples (uniform molecular orientation) compared with the spin-coated samples (molecular orientation varied significantly). TERS investigation of a 2DP constructed from a monomer containing three fluoro-substituted anthracene blades and a bridgehead carboxylic acid was performed by Müller et al.42 The bridgehead C–C stretching mode at 970 cm−1 was used as the Raman marker for tetrafluoroanthracene dimers. TERS measurements confirmed a successful 2D polymerisation, albeit some unreacted monomers were also detected and a reaction conversion of about 90% was estimated.
The photoinduced polymerisation process of macrocyclic anthracene monomers into 2D covalent sheets was investigated using TERS by Zheng et al.43 The success of the polymerisation was verified using TERS and in situ fluorescence decay experiments. DFT calculations showed that the Raman signal at 1385 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching mode of anthracene) disappeared in the spectrum of a fully polymerised model, whilst the 995 cm−1 signal (ring breathing mode of cyclophane) was present in both the monomer and polymer models. Therefore, the intensity ratio of 1385 cm−1 and 995 cm−1 Raman signals (I1385/I995) was used to track polymerisation conversion. The I1385/I995 ratio indicated an increase in the polymerisation conversion up to 6.5 h of UV irradiation. Furthermore, a high degree of crystallinity was confirmed in the final product, qualifying it to be a 2DP.
C stretching mode of anthracene) disappeared in the spectrum of a fully polymerised model, whilst the 995 cm−1 signal (ring breathing mode of cyclophane) was present in both the monomer and polymer models. Therefore, the intensity ratio of 1385 cm−1 and 995 cm−1 Raman signals (I1385/I995) was used to track polymerisation conversion. The I1385/I995 ratio indicated an increase in the polymerisation conversion up to 6.5 h of UV irradiation. Furthermore, a high degree of crystallinity was confirmed in the final product, qualifying it to be a 2DP.
Homo- and co-polymerisation of two different amphiphilic monomers containing a rigid triptycene core and three anthracene blades were examined using TERS by Wang et al.44 In addition to proving the polymerisation, the conversion number and the degree of crystallinity were estimated to be >85% and >90%, respectively. In the case of 93% conversion, coverage of the fully ordered monolayer covalent sheets was estimated to be 710 nm2, which was interspersed by small areas (2.8%) of the unreacted monomers. The study demonstrated that TERS could be used to examine the long-range order in 2DPs.
The above-mentioned examples demonstrate the ability of TERS to provide novel scientific insights in 2DPs, e.g., determine the polymerisation mechanism, polymerisation conversion rate, degree of crystallinity, molecular orientation, and presence of nanodefects. This information is not only essential for the fundamental understanding but also for the measurement of polymer properties that should be tuned for bespoke applications.
Although TERS has shown great promise for nanoscale investigation of the 2DPs, in the future, it would be beneficial to employ additional complementary techniques to support the characterisation of molecular orientation in these systems. For example, high-resolution STM, polarised AFM–IR,45 or even low-resolution techniques, such as subtractive normalised interfacial Fourier transform infrared spectroscopy,46 polarisation-modulation infrared reflection-absorption spectroscopy,47 and angle-dependent XPS,48 could be used as complementary tools to confirm the average molecular orientation of the unreacted monomers and 2DPs, which would further enhance the reliability of TERS for nanoanalysis of these novel 2D molecular materials.
3. Biomimetic lipid membranes
The first TERS study of a biomimetic lipid membrane was performed by Böhme et al. on a mica-supported lipid bilayer composed of palmitoyloleoylphosphatidylcholine (POPC)/dioleoylphosphatidylserine (DOPS).49 The AFM topography image and three TERS spectra measured consecutively from one location on the lipid bilayer are shown in Fig. 3a and b, respectively. The TERS spectra showed a high signal-to-noise ratio but differed significantly in terms of the number, position, and relative intensity of Raman bands, which was attributed to the dynamic behaviour of the lipid structure and TERS surface selection rules. In another study, Böhme et al.50 investigated a supported lipid bilayer of 1,2-dioleoy-sn-glycero-3-phosphocholine (DOPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) labelled with streptavidin using TERS. Using a clean metal-coated probe, TERS measurements were performed along two lines across the lipid bilayer, as shown in Fig. 3c. Like the previous study, the TERS spectra differed in terms of the number, position, and relative intensity of Raman bands. Using the literature-based assignment of Raman bands, the authors classified the TERS spectra into three types: containing typical (1) lipid bands, (2) protein bands or (3) both lipid and protein bands, as shown in Fig. 3d. Based on this classification, the distribution of lipid and protein molecules along the two lines measured on the bilayer surface was proposed (Fig. 3c).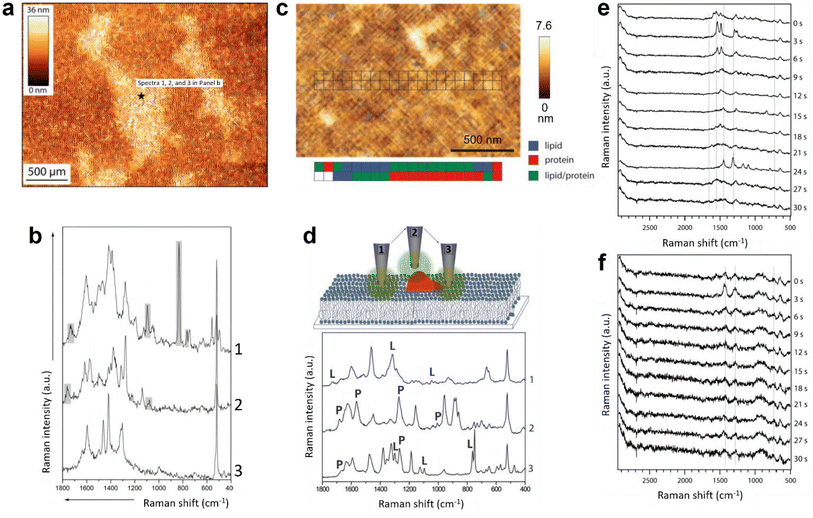 | ||
| Fig. 3 (a) AFM topography image of a POPC/DOPS bilayer on mica. TERS spectra were recorded at the marked spot (black star). (b) TERS spectra measured consecutively at the spot marked in panel a. Panels a and b are adapted from ref. 49 with permission from John Wiley & Sons, copyright 2009. (c) AFM topography image of a streptavidin-labelled phospholipid film. The positions where TERS spectra were recorded are marked. A false-colour map of the lipid and protein regions is shown below the topography image. (d) Schematic diagram illustrating different tip–sample interactions during the TERS measurement of the streptavidin-labelled phospholipid film and a plot of the corresponding TERS spectra. Marker bands for lipids and proteins are labelled with L and P, respectively. Panels c and d are adapted from ref. 50 with permission from John Wiley & Sons, copyright 2010. Time-series TERS spectra of supported (e) DOPC and (f) DPPC bilayers. Panels e and f are adapted from ref. 51. | ||
A major issue to study biomembranes in their native state using AFM–TERS is probe degradation due to the rapid delamination of the plasmonic coating in the aqueous environment.52,53 The first in situ AFM–TERS measurement of DOPC and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) lipid bilayers was attempted by Nakata et al.51 using alumina- and silver-coated tungsten tips. Supported lipid bilayers were deposited on glass using the vesicle deposition technique. Time-series TERS spectra of the DOPC bilayer showed a large spectral fluctuation (Fig. 3e), whereas the TERS spectra of the DPPC bilayer were found to be relatively stable (Fig. 3f). Although the signal-to-noise ratio of the measured TERS spectra was not very high, this study showed the potential of TERS to characterise inhomogeneities and/or diffusion of the phospholipid bilayers in an aqueous environment.
All TERS studies of the supported lipid layers discussed so far focused on the fingerprint region of the Raman spectrum. In fact, the C–H stretching region (2800–3000 cm−1) of the lipid membranes also contains valuable information regarding the orientation, ordering, and protein–lipid interaction.54 Our group utilised Raman signals in the C–H and C–D stretching regions to investigate a mixed d62-DPPC/DOPC monolayer prepared via LB method on a template-stripped Au (TS-Au) substrate.55 Large area (128 × 128 pixels) high-resolution STM–TERS imaging of the d62-DPPC/DOPC lipid monolayer was performed with a step size of 47 nm. Using the intensity ratio of the Raman bands at 2100 cm−1 (C–D stretching vibration, ICD) and 2900 cm−1 (C–H stretching vibration, ICH), phase-separated d62-DPPC regions could be visualised in a mixed lipid monolayer, as shown in Fig. 4a. Notably, no Raman signal was observed in the fingerprint region from the lipids (Fig. 4b). In a different study, we performed AFM–TERS measurement of a mixed monolayer of DPPC and 15-hexadecynoic acid (PA-15-yne) on mica. Interestingly, in the fingerprint region, the TERS and far-field Raman spectra only exhibited Raman bands of the mica substrate and no bands from the DPPC/PA-15-yne monolayer were observed. However, a strong plasmonic enhancement in the C–H stretching region was observed in the near-field (TERS spectrum) as shown in Fig. 4c, which indicates that the C–H stretching region can be signifcant for the TERS analysis of lipid membranes.
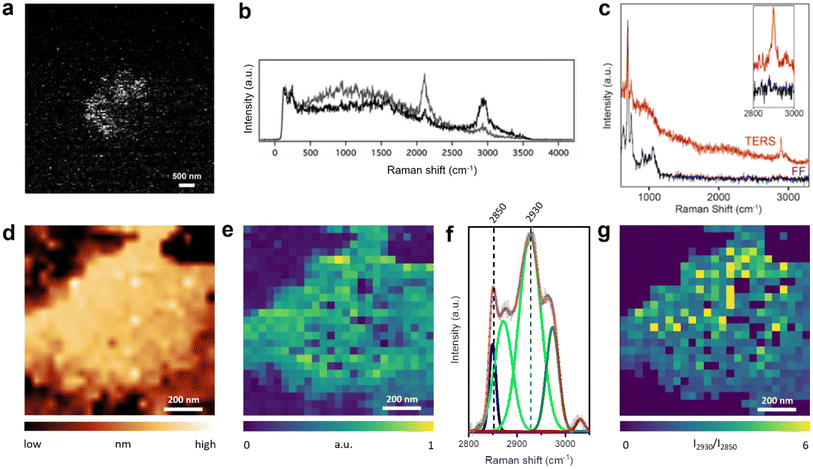 | ||
| Fig. 4 (a) TERS image of the ICD/ICH ratio of a d62-DPPC/DOPC mixed monolayer. (b) Representative Raman spectra from dark (black) and bright (grey) pixels in panel a. Panels a and b are adapted from ref. 55 (c) TERS and far-field Raman spectra of a mixed DPPC/PA-15-yne monolayer on mica. The Raman bands in the 600–1100 cm−1 region come from the mica substrate. The inset shows an enlarged image of the C–H stretching region. (d) STM topography image and (e) TERS image constructed using the intensity of the C–H stretching region (2800–3000 cm−1) of a DPPC monolayer on Au(111). (f) Average TERS spectrum of the DPPC monolayer region shown in panel e, deconvoluted using fitted Gaussian curves. (g) TERS image of the DPPC monolayer constructed using the I2930/I2850 ratio. Panels d–g are adapted from ref. 31 with permission from John Wiley & Sons, copyright 2021. | ||
More recently, we reported corelative STM and TERS imaging of a DPPC monolayer supported on an Au(111) surface as shown in Fig. 4d and e.31 Reproducible high-resolution TERS imaging was achieved using the C–H stretching Raman signals with a step size of 20–40 nm. A high signal-to-noise ratio was achieved in the TERS spectra, which allowed a detailed investigation of the lipid monolayer via deconvolution of the Raman bands in the C–H stretching region, as shown in Fig. 4f. Using the intensity ratio of the 2930 cm−1 band (I2930, Fermi resonance associated with terminal methyl symmetric stretching and C–H bending modes) and the 2850 cm−1 band (I2850, methylene symmetric stretching), we were able to map molecular disorder in the DPPC monolayer at the nanoscale, as shown in Fig. 4g.56 Furthermore, TERS imaging revealed the presence of 40–120 nm size holes in the lipid film. This study showed that a DPPC monolayer transferred at high surface pressure can exhibit significant heterogeneities at the nanoscale.
The aforementioned studies confirm that TERS has developed into a powerful and reliable nanoscale characterisation tool to study supported lipid membranes. From the single-point measurements in an ambient environment to the time-resolved measurements in liquids, the capability of AFM–TERS to reveal the nanoscale biophysical phenomenon without exogenous labelling is advancing rapidly. Similarly, STM–TERS of model lipid monolayer systems has also progressed significantly, from imaging the lipid distribution in a raft-like system to deciphering the molecular orientation in a supported lipid monolayer. These studies of biomimetic systems are expected to lay the groundwork for understanding more complex biological systems like cell membranes using TERS.
Notably, several TERS studies of biomimetic systems utilise a mica substrate in non-gap mode. To achieve a high signal-to-noise ratio in non-gap mode TERS measurements, some key criteria need to be met. For example, the TERS probe should have a uniform metal coating with the presence of corrugated nanostructures at the apex. The excitation and collection optical pathways of the TERS system should be perfectly aligned. The experimental parameters including the LSPR resonant excitation laser, laser power, and spectral acquisition time should be carefully optimised without photodamaging the sample. Furthermore, the instrument/sample drift is a significant issue in hyperspectral TERS imaging, as highlighted in recent works.32,57 Therefore, the step size of TERS imaging should be selected carefully depending on the size of the region of interest. For TERS measurements of homogeneous samples such as uniform supported lipid monolayers or bilayers, a large step size (up to 50 nm) can be an effective method to obtain an overview of the sample. However, for a thorough analysis of the local nanoscale features such as defects, molecular orientation, etc., a small step size of a few nm is desirable.
4. Biological cell membranes
The cell membrane is a protein-rich lipid bilayer that acts as a 2D semi-permeable barrier that regulates molecular traffic across the cell. It also provides a platform for important cellular functions like cell proliferation, adhesion, and migration, which is why nanoscopic membrane components are the targets of upto 45% of the therapeutic drugs under development.58 The first TERS measurement of a cell membrane was performed on an S. epidermidis cell by Neugebauer et al.59 Three TERS spectra were measured from different locations on the bacterial surface and a significant variation in the number, position, and relative intensity of Raman bands was observed. On the assumption that spectral fluctuations came from the chemical heterogeneity and/or dynamics of the bacterial membrane and not from any carbonaceous/organic contaminants, the authors assigned the observed Raman bands to proteins, lipids or saccharides using SERS literature. TERS investigation of a human dermal-derived keratinocyte surface was performed by Böhme et al.49 TERS probe contamination was ruled out with a control measurement, but only three TERS spectra were measured, which, like in the previous study, showed significant spectral variability. The authors interpreted some of the Raman bands by invoking the strong dependence of TERS enhancement on the probe–sample distance. For example, in the case of lipids, Raman bands at 780, 810, 1095, and 1210 cm−1 were assigned to the vibrations of the phosphate groups presumably because of the proximity of TERS tip to the lipid head.The first successful attempt to characterise the antibody–antigen interaction on a cell membrane surface was carried out by the Schultz group.60 Hyperspectral TERS imaging was performed on anti-IgG labelled Au nanoparticles (NPs) bound to the plasma membrane of colon cancer cells. NPs were identified through the Raman marker bands of anti-IgG. Isolated Au NPs showed TERS spectra containing Raman bands that matched with the reference Raman spectra of anti-IgG (1170, 1210, 1340, 1465, and 1485 cm−1) and the antibody–antigen interaction (485 cm−1). In contrast, TERS spectra of the aggregated Au NPs were found to be significantly more complex containing Raman bands from anti-IgG as well as other molecules. The complexity of the aggregated Au NPs in the TERS spectra was attributed to the presence of multiple “hotspots” capturing molecules in different orientations and locations and/or the occurrence of “spurious” signals from the atmospheric carbon contaminants (ACCs). TERS characterisation of the membrane of cyanobacteria, Halobacterium salinarum S9 (HSS9), was performed by Deckert-Gaudig et al.61 Because of its colour, this type of membrane is commonly referred to as a purple membrane (PM). The bacterial membrane of HSS9 is highly rich in a light-sensitive proton pump protein called bacteriorhodopsin (bR). Using the resonantly enhanced retinal moiety of bR, the authors tried to identify PM patches on the HSS9 surface. From the TERS measurement of the isolated PM patches, the authors first identified the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH+ stretching modes at 1540 and 1635 cm−1, respectively, as unique Raman markers of the retinal moiety. Using these marker bands, the distribution of the PM on the HS9 membrane was mapped with a step size of 75 nm in a 300 × 375 nm2 area. However, as in some of the aforementioned studies, within the PM patches significant variations in the position, number, and relative intensity of Raman bands were observed, as shown in Fig. 5a–c. In such situations, an unequivocal interpretation of the TERS spectra can be difficult, considering the common observation of spurious signals (attributed to the ACCs) in ambient TERS measurements as shown in Fig. 5d, where multiple Raman bands that do not belong to the analyte are present.63
NH+ stretching modes at 1540 and 1635 cm−1, respectively, as unique Raman markers of the retinal moiety. Using these marker bands, the distribution of the PM on the HS9 membrane was mapped with a step size of 75 nm in a 300 × 375 nm2 area. However, as in some of the aforementioned studies, within the PM patches significant variations in the position, number, and relative intensity of Raman bands were observed, as shown in Fig. 5a–c. In such situations, an unequivocal interpretation of the TERS spectra can be difficult, considering the common observation of spurious signals (attributed to the ACCs) in ambient TERS measurements as shown in Fig. 5d, where multiple Raman bands that do not belong to the analyte are present.63
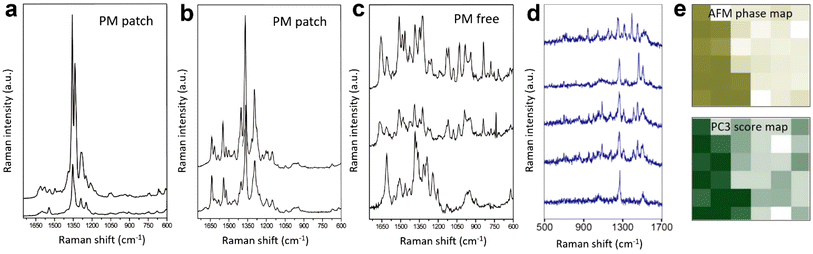 | ||
| Fig. 5 TERS spectra measured at (a) a PM patch, (b) another PM patch, and (c) a PM-free patch. Panels a–c are adapted from ref. 61 with permission from John Wiley & Sons, copyright 2012. (d) Examples of spurious TERS spectra from carbonaceous/organic contaminants measured from a self-assembled monolayer (SAM) of 2-chloro-4-nitrobenzene-1-thiol on Au. (e) Top: AFM phase image of a Bacillus subtilis spore. Bottom: PC3 scores map of the TERS spectra measured in the same area. PC3 is not assigned to a specific class of compounds, nonetheless, there is a clear correlation between the AFM and TERS images. Panel e is adapted from ref. 62. | ||
The outer layer of the Bacillus subtilis spore was analysed using TERS by Rusciano et al.62 As in the case of previous studies, individual spectra measured in the TERS image showed a significant point-to-point variation. However, the authors demonstrated that although no correlation could be established between the AFM phase image and the TERS image of individual Raman signals, principal component analysis (PCA) of the hyperspectral TERS data provided a clear correlation. Particularly, PC2 and PC3 score maps matched very well the AFM phase image of a ridge feature on the spore surface, as shown in Fig. 5e. Even though PCA components are difficult to assign to a specific chemical element or class of compounds due to the inherent complexity of the bacterial surface, this work highlighted the potential of higher-level data analysis to extract meaningful information from the TERS measurements of complex specimens such as biological cell membranes.
Conceivably, TERS measurements of biomembranes are relatively more complex than the simpler, well-defined, biomimetic systems. Nevertheless, the presented studies clearly demonstrate the potential of TERS to detect various cell membrane components and provide information about their structural integrity, mutual interaction, and spatial distribution. Such molecular insights can enable a deeper understanding of fundamental cellular processes including cell growth, differentiation, signal transduction, molecular transportation, cellular communication with its environment, etc.64 Furthermore, TERS has the ability to validate the lipid raft theory, which is still a debatable concept in the biological research community.65
As in the case of biomimetic systems, the TERS studies on cell membranes discussed above primarily focused on the fingerprint region of the Raman spectrum. However, as demonstrated by Pandey et al.,31 the C–H stretching region also provides rich molecular information for the structural and chemical analysis of lipid membranes at the nanoscale, which future TERS studies of biological cell membranes should exploit.
5. 2D reactive systems
Bottom-up fabrication of surface nanostructures via different combinations and spatial arrangements of building blocks provides 2D reactive systems suitable for developing functional devices with controllable properties.66–69 For example, organometallic structures such as metalloporphyrins (MPs) and metallophthalocyanines (MPCs) have attracted a lot of interest because of their remarkable catalytic properties and application in the biological and chemical systems.70–74 However, the structure–reactivity relationships in these materials are poorly understood at the nanoscale due to the lack of information related to molecular specificity, configuration, and orientation. TERS is well suited to tackle these challenges since it can be applied for nanoscale chemical characterisation of interfaces under different environmental conditions.18,28,53,75–77A lot of effort has been applied to achieving high-resolution visualisation of individual MP/MPC molecules using TERS by several research groups.78–83 A pioneering study was conducted by the Dong group, in which the first TERS image of a single porphyrin molecule (H2TBPP) resolving its inner structure with a spatial resolution of <1 nm were obtained.83 Several years later, the Apkarian group resolved the vibrational normal modes of a cobalt–tetraphenyl porphyrin (CoTPP) molecule at the atomic level using TERS under cryogenic ultrahigh vacuum conditions.78 Besides the high-resolution chemical imaging, TERS was also successfully utilised to resolve the conformation,82–84 orientation,80,81 and arrangement85 of 2D reactive systems. For example, the Pettinger group probed molecular ordering within a SAM of CoTPP on Au(111) using TERS.85 Later on, the Van Duyne group used TERS to detect the conformational difference between neighbouring H2TBPP molecules on a Cu(111) surface.84
Recently, our group also applied TERS to investigate the molecular orientation of a CoTPP–4-mercaptopyridine (4PySH) coordination system (schematically illustrated in Fig. 6a) immobilised on a Au surface with a spatial resolution of ca. 2 nm.86Fig. 6b shows three typical TERS spectra of the CoTPP–4PySH complex, extracted from a high-resolution TERS image on an atomically-flat Au(111) surface. The signal intensity of the bands highlighted by red stripes varied from type A to type C, which suggests that the vibrational modes corresponding to these bands are plasmonically enhanced to a different extent. Based on the TERS surface selection rules81,83 and theoretical simulations87 of 4PySH molecule adsorbed on an Au(111) surface, we proposed that these spectral differences reflect different orientations of the coordinated CoTPP–4PySH species. Fig. 6c shows the DFT-calculated Raman spectra of the CoTPP 4PySH coordination complex with different tilt angles. By correlating the measured TERS spectra with DFT simulations, the molecular orientation of CoTPP in the CoTPP–4PySH coordination complex on Au(111) could be determined.
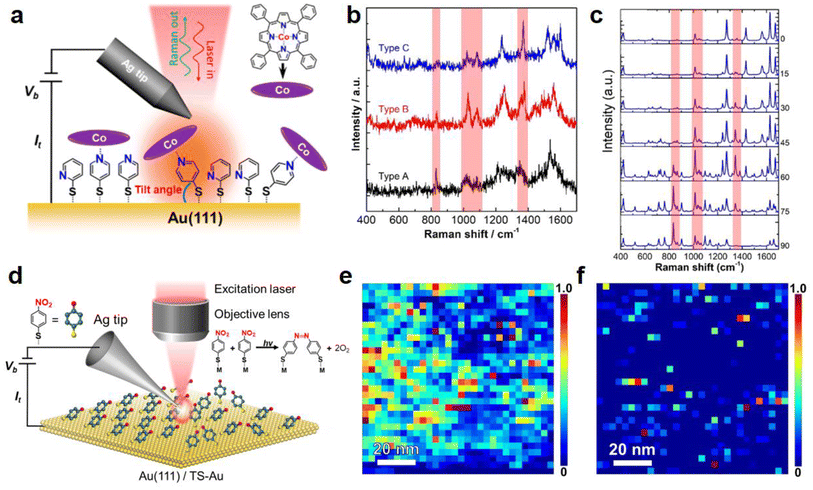 | ||
| Fig. 6 (a) Schematic diagram illustrating the STM–TERS setup to probe the CoTPP species on a TS-Au surface modified with a 2PySH and 4PySH mixed adlayer. (b) Three typical TERS spectra showing variable peak intensities of the Raman bands highlighted by red stripes. (c) DFT-calculated Raman spectra of a CoTPP–4PySH–Au2 complex with tilt angles from 0°–90° showing significant spectral variations. Panels a–c are adapted from ref. 86 with permission from the American Chemical Society, copyright 2021. (d) Schematic diagram of an STM–TERS setup used to investigate the role of molecular arrangement in the photocatalytic conversion of 4-NTP → DMAB on polycrystalline and single-crystal Au surfaces. (e and f) High-resolution TERS images of DMAB formation on samples prepared via drop-cast and immersion protocols on a polycrystalline TS-Au surface. Panels d–f are adapted from ref. 29 with permission from the American Chemical Society, copyright 2022. | ||
Plasmon-driven photocatalytic coupling reactions have been used as model systems in SERS and TERS research for several decades.88 With the assistance of hot electrons generated in the near-field, 4-aminothiophenol (4-ATP) and 4-nitrobenzenethiol (4-NTP) can be converted to p,p′-dimercaptoazobenzene (DMAB) on plasmonic metal surfaces.88–90 It has been shown that the efficiency of these plasmon-driven photocatalytic coupling reactions is strongly influenced by the temperature,91 excitation laser,92 substrate,93,94 and surrounding environment.95,96 However, a clear structure–reactivity relationship for the nanoscale arrangement of the reactant species on the catalyst surface has not been established. In this context, Sun et al. investigated the effect of adsorption orientation on the photocatalytic conversion of 4-NTP and 4-ATP → DMAB on single-crystal Ag and Au surfaces using TERS.94 However, the results lacked support from direct visualisation of the arrangement of 4-NTP and 4-ATP molecules adsorbed on the single crystal surfaces.
Our group used a combination of TERS, STM, and DFT simulations to investigate the role of the molecular arrangement in the photocatalytic coupling of 4-NTP → DMAB on single-crystal and polycrystalline Au surfaces, as schematically illustrated in Fig. 6d.29 Two samples were investigated: the first sample was formed via the “dropcast” protocol, where a drop of ethanolic 4-NTP solution was placed on the Au surface and allowed to evaporate in <1 min. The second sample was prepared via the “immersion” protocol, where 4-NTP molecules were allowed to self-assemble on a Au surface by immersing it in an ethanolic 4-NTP solution for >12 h. The idea was that the dropcast protocol would lead to a disordered adlayer, while the immersion protocol would allow the formation of a well-ordered SAM. Interestingly, high-resolution TERS imaging of the 4-NTP → DMAB reaction on an Au surface showed a significantly higher level of DMAB product formation on the drop-cast sample (Fig. 6e) compared with the immersion sample (Fig. 6f).
To understand this behaviour, we performed molecular-resolution STM imaging, which revealed that only 8% of the drop-cast sample surface was covered with well-ordered molecular domains; on the other hand, 76% of the immersion sample surface contained a well-ordered molecular adlayer. Despite the difference in molecular arrangement, the intermolecular distance in the ordered domains of both samples was found to be ca. 0.8 nm. DFT simulations based on these intermolecular distance and surface coverage parameters revealed that the molecules in the well-ordered domains were adsorbed at a 69° angle with the vertical meaning they were lying almost flat on the substrate. Furthermore, the energy required for a molecule to rotate around the vertical axis was found to be 0.3 eV, which is readily available under ambient conditions. This meant that in the close-packed arrangement, the –NO2 head groups of the adjacent 4-NTP molecules could never meet each other via rotation around the vertical axis, providing the physical rationale for its lower 4-NTP → DMAB conversion efficiency. However, a favourable configuration for 4-NTP → DMAB conversion could be more easily adopted in the disordered domains of the drop-cast sample, leading to a significantly higher reaction efficiency. This fundamental study established the first direct structure–reactivity correlation in the photocatalytic coupling of 4-NTP → DMAB and highlighted the critical role of the molecular arrangement on the efficiency of on-surface coupling reactions in heterogeneous catalysis at large.
Despite the unique advantages of high sensitivity and nanoscale spatial resolution, the applications of TERS to study on-surface chemical transformations as in heterogenous catalysis, molecular degradation and co-coordination chemistry are still in their infancy. However, with significant improvements in the yield,97,98 lifetime,28,99 and chemical inertness28,100 of TERS probes, rapid progress in this area is envisaged.
6. Conclusions and future outlook
The studies discussed in this review have shown that TERS can yield valuable information on different 2D molecular materials of chemical and biological interest such as 2D polymers, biomimetic lipid membranes, cellular membranes, and reactive monolayers. The particular strength of TERS is that spectroscopic information is available with a lateral resolution of only a few nm under ambient conditions, without exogenous labelling and destroying the sample. Particularly novel scientific insights were obtained for cases where hyperspectral TERS imaging was combined with complementary data from other techniques, e.g., molecular-resolution STM images of the same sample, as demonstrated by Cai et al.29Despite the obvious advantages of TERS, there are still some hurdles to overcome, as well as some control and complementary measurements that should be routinely performed, particularly when studying complex samples: the correlation of high-resolution topographic (AFM or STM) imaging with TERS should be carried out whenever possible. This is not always easy; for example, lipid bilayers are too thick to electrons to tunnel through, i.e., STM and gap-mode TERS can not be applied in this case. In AFM–TERS, it should be carefully considered whether contact mode or tapping mode is used to perform such correlative imaging.63,101 The interpretation of a mismatch between high-resolution topographic imaging and TERS is equally important.
The high sensitivity, molecular specificity, and spatial resolution of hyperspectral TERS nanoanalysis can potentially address many unanswered scientific questions regarding “soft” 2D materials. For example, in the context of cellular membranes, TERS could be used to study structural variations between different cell types, which can be used as potential markers to distinguish them. However, in the TERS studies of biomimetic and biological systems, it is essential to have appropriate reference data and control measurements. For assigning TERS bands, Raman spectra of the reference compounds obtained in the same experimental setup are important in our opinion and preferable to the assignment based on the literature. It should be always demonstrated that an actual TERS enhancement exists, and tip contamination and sample decomposition must be ruled out by control experiments.
In biological samples, the information depth of TERS needs to be understood better. Currently, there are inconsistent reports in the literature – some papers claim that the Raman signal of compounds tens of nm away from the TERS tip can be enhanced102 and others claim that only molecular residues virtually in contact with the TERS tips would experience an enhancement of Raman signals.49 Nevertheless, we see great promise in continuing to study such systems using TERS. In particular, the knowledge of biomimetic systems will set a solid basis for understanding more complex and relevant biological systems like cellular membranes. Last but not least, since TERS can be performed in a liquid environment,53 2D molecular systems such as lipid monolayers, bilayers, and natural membranes should more often be studied in their natural environment, i.e. in liquid or buffer, even if this adds an additional layer of complication to the TERS experiments.
Author contributions
All authors contributed equally to the writing of this article.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support from the European Union through ERC grant no. 741431 (2D Nano-Spec).References
- A. P. Côté, A. I. Benin, N. W. Ockwig, M. O'Keeffe, A. J. Matzger and O. M. Yaghi, Science, 2005, 310, 1166–1170 CrossRef PubMed.
- P. Payamyar, B. T. King, H. C. Öttinger and A. D. Schlüter, Chem. Commun., 2016, 52, 18–34 RSC.
- D. J. Murray, D. D. Patterson, P. Payamyar, R. Bhola, W. Song, M. Lackinger, A. D. Schlüter and B. T. King, J. Am. Chem. Soc., 2015, 137, 3450–3453 CrossRef CAS PubMed.
- W. Liu, X. Luo, Y. Bao, Y. P. Liu, G.-H. Ning, I. Abdelwahab, L. Li, C. T. Nai, Z. G. Hu, D. Zhao, B. Liu, S. Y. Quek and K. P. Loh, Nat. Chem., 2017, 9, 563–570 CrossRef CAS PubMed.
- Y. Zhao, R. H. M. Bernitzky, M. J. Kory, G. Hofer, J. Hofkens and A. D. Schlüter, J. Am. Chem. Soc., 2016, 138, 8976–8981 CrossRef CAS PubMed.
- H. Sahabudeen, H. Qi, B. A. Glatz, D. Tranca, R. Dong, Y. Hou, T. Zhang, C. Kuttner, T. Lehnert, G. Seifert, U. Kaiser, A. Fery, Z. Zheng and X. Feng, Nat. Commun., 2016, 7, 1–8 Search PubMed.
- T. Humplik, J. Lee, S. C. O'Hern, B. A. Fellman, M. A. Baig, S. F. Hassan, M. A. Atieh, F. Rahman, T. Laoui, R. Karnik and E. N. Wang, Nanotechnology, 2011, 22, 292001 CrossRef CAS PubMed.
- Y. Saylan, Ö. Erdem, F. Inci and A. Denizli, Biomimetics, 2020, 5, 20 CrossRef CAS PubMed.
- E. T. Castellana and P. S. Cremer, Surf. Sci. Rep., 2006, 61, 429–444 CrossRef CAS PubMed.
- V. V. Korolkov, A. Summerfield, A. Murphy, D. B. Amabilino, K. Watanabe, T. Taniguchi and P. H. Beton, Nat. Commun., 2019, 10, 1537 CrossRef PubMed.
- A. N. Chaika, in Surface Science Tools for Nanomaterials Characterisation, Springer Berlin Heidelberg, Berlin, Heidelberg, 2015, pp. 561–619 Search PubMed.
- P. Lasch and D. Naumann, Biochim. Biophys. Acta, Biomembr., 2006, 1758, 814–829 CrossRef CAS PubMed.
- R. H. Webb, Rep. Prog. Phys., 1996, 59, 427–471 CrossRef.
- J. A. Whitby, F. Östlund, P. Horvath, M. Gabureac, J. L. Riesterer, I. Utke, M. Hohl, L. Sedláček, J. Jiruše, V. Friedli, M. Bechelany and J. Michler, Adv. Mater. Sci. Eng., 2012, 2012, 1–13 CrossRef.
- L. Schermelleh, A. Ferrand, T. Huser, C. Eggeling, M. Sauer, O. Biehlmaier and G. P. C. Drummen, Nat. Cell Biol., 2019, 21, 72–84 CrossRef CAS PubMed.
- D. Kurouski, A. Dazzi, R. Zenobi and A. Centrone, Chem. Soc. Rev., 2020, 49, 3315–3347 RSC.
- N. Kumar, S. Mignuzzi, W. Su and D. Roy, EPJ Tech. Instrum., 2015, 2, 9 CrossRef.
- N. Kumar, B. M. Weckhuysen, A. J. Wain and A. J. Pollard, Nat. Protoc., 2019, 14, 1169–1193 CrossRef CAS PubMed.
- P. Verma, Chem. Rev., 2017, 117, 6447–6466 CrossRef CAS PubMed.
- W. Su, N. Kumar, S. Mignuzzi, J. Crain and D. Roy, Nanoscale, 2016, 8, 10564–10569 RSC.
- W. Su, N. Kumar, A. Krayev and M. Chaigneau, Nat. Commun., 2018, 9, 2891 CrossRef PubMed.
- W. Su, A. Esfandiar, O. Lancry, J. Shao, N. Kumar and M. Chaigneau, Chem. Commun., 2021, 57, 6895–6898 RSC.
- W. Su, N. Kumar, H. Shu, O. Lancry and M. Chaigneau, J. Phys. Chem. C, 2021, 125, 26883–26891 CrossRef CAS.
- N. Kumar, S. Marchesini, T. Howe, L. Edwards, B. Brennan and A. J. Pollard, J. Chem. Phys., 2020, 153, 184708 CrossRef CAS PubMed.
- Z.-F. Cai, T. Käser, N. Kumar and R. Zenobi, J. Phys. Chem. Lett., 2022, 13, 4864–4870 CrossRef CAS PubMed.
- D. Mrđenović, D. Abbott, V. Mougel, W. Su, N. Kumar and R. Zenobi, ACS Appl. Mater. Interfaces, 2022, 14, 24938–24945 CrossRef PubMed.
- N. Kumar, B. Stephanidis, R. Zenobi, A. J. Wain and D. Roy, Nanoscale, 2015, 7, 7133–7137 RSC.
- N. Kumar, C. S. Wondergem, A. J. Wain and B. M. Weckhuysen, J. Phys. Chem. Lett., 2019, 10, 1669–1675 CrossRef CAS PubMed.
- Z. F. Cai, J. P. Merino, W. Fang, N. Kumar, J. O. Richardson, S. De Feyter and R. Zenobi, J. Am. Chem. Soc., 2022, 144, 538–546 CrossRef CAS PubMed.
- N. Kumar, A. Zoladek-Lemanczyk, A. A. Y. Guilbert, W. Su, S. M. Tuladhar, T. Kirchartz, B. C. Schroeder, I. McCulloch, J. Nelson, D. Roy and F. A. Castro, Nanoscale, 2017, 9, 2723–2731 RSC.
- Y. Pandey, N. Kumar, G. Goubert and R. Zenobi, Angew. Chem., Int. Ed., 2021, 60, 19041–19046 CrossRef CAS PubMed.
- D. Mrđenović, W. Ge, N. Kumar and R. Zenobi, Angew. Chem., Int. Ed., 2022, 61(43), e202210288 CrossRef PubMed.
- N. Kumar, M. M. Drozdz, H. Jiang, D. M. Santos and D. J. Vaux, Chem. Commun., 2017, 53, 2451–2454 RSC.
- R. J. C. Brown and M. J. T. Milton, J. Raman Spectrosc., 2008, 39, 1313–1326 CrossRef CAS.
- S. L. Kleinman, R. R. Frontiera, A. I. Henry, J. A. Dieringer and R. P. Van Duyne, Phys. Chem. Chem. Phys., 2013, 15, 21–36 RSC.
- B. Sharma, R. R. Frontiera, A. I. Henry, E. Ringe and R. P. Van Duyne, Mater. Today, 2012, 15, 16–25 CrossRef CAS.
- G. M. Lerman and U. Levy, Opt. Express, 2008, 16, 4567 CrossRef PubMed.
- L. Opilik, P. Payamyar, J. Szczerbiński, A. P. Schütz, M. Servalli, T. Hungerland, A. D. Schlüter and R. Zenobi, ACS Nano, 2015, 9, 4252–4259 CrossRef CAS PubMed.
- W. Dai, F. Shao, J. Szczerbiński, R. McCaffrey, R. Zenobi, Y. Jin, A. D. Schlüter and W. Zhang, Angew. Chem., Int. Ed., 2016, 55, 213–217 CrossRef CAS PubMed.
- F. Shao, W. Dai, Y. Zhang, W. Zhang, A. D. Schlüter and R. Zenobi, ACS Nano, 2018, 12, 5021–5029 CrossRef CAS PubMed.
- F. Shao, V. Müller, Y. Zhang, A. D. Schlüter and R. Zenobi, Angew. Chem., Int. Ed., 2017, 56, 9361–9366 CrossRef CAS PubMed.
- V. Müller, F. Shao, M. Baljozovic, M. Moradi, Y. Zhang, T. Jung, W. B. Thompson, B. T. King, R. Zenobi and A. D. Schlüter, Angew. Chem., Int. Ed., 2017, 56, 15262–15266 CrossRef PubMed.
- L.-Q. Zheng, M. Servalli, A. D. Schlüter and R. Zenobi, Chem. Sci., 2019, 10, 9673–9678 RSC.
- W. Wang, F. Shao, M. Kröger, R. Zenobi and A. D. Schlüter, J. Am. Chem. Soc., 2019, 141, 9867–9871 CrossRef CAS PubMed.
- Z. Wang, B. Sun, X. Lu, C. Wang and Z. Su, Macromolecules, 2019, 52, 9639–9645 CrossRef CAS.
- V. Zamlynny and J. Lipkowski, in Advances in Electrochemical Sciences and Engineering, 2006, vol. 9, pp. 315–376 Search PubMed.
- A. H. Kycia, Z. Su, C. L. Brosseau and J. Lipkowski, in Vibrational Spectroscopy at Electrified Interfaces, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2013, pp. 345–417 Search PubMed.
- M. C. Burrell, M. D. Butts, D. Derr, S. Genovese and R. J. Perry, Appl. Surf. Sci., 2004, 227, 1–6 CrossRef CAS.
- R. Böhme, M. Richter, D. Cialla, P. Rösch, V. Deckert and J. Popp, J. Raman Spectrosc., 2009, 40, 1452–1457 CrossRef.
- R. Böhme, D. Cialla, M. Richter, P. Rösch, J. Popp and V. Deckert, J. Biophotonics, 2010, 3, 455–461 CrossRef PubMed.
- A. Nakata, T. Nomoto, T. Toyota and M. Fujinami, Anal. Sci., 2013, 29, 865–869 CrossRef CAS PubMed.
- T. Schmid, B. S. Yeo, G. Leong, J. Stadler and R. Zenobi, J. Raman Spectrosc., 2009, 40, 1392–1399 CrossRef CAS.
- N. Kumar, W. Su, M. Veselý, B. M. Weckhuysen, A. J. Pollard and A. J. Wain, Nanoscale, 2018, 10, 1815–1824 RSC.
- K. Larsson and R. P. Rand, Biochim. Biophys. Acta, Lipids Lipid Metab., 1973, 326, 245–255 CrossRef CAS PubMed.
- L. Opilik, T. Bauer, T. Schmid, J. Stadler and R. Zenobi, Phys. Chem. Chem. Phys., 2011, 13, 9978 RSC.
- C. B. Fox, R. H. Uibel and J. M. Harris, J. Phys. Chem. B, 2007, 111, 11428–11436 CrossRef CAS PubMed.
- R. Kato, T. Moriyama, T. Umakoshi, T. Yano and P. Verma, Sci. Adv., 2022, 8, 1–11 Search PubMed.
- A. B. van Spriel, G. van den Bogaart and A. Cambi, Front. Physiol., 2015, 6, 322–330 Search PubMed.
- U. Neugebauer, P. Rösch, M. Schmitt, J. Popp, C. Julien, A. Rasmussen, C. Budich and V. Deckert, ChemPhysChem, 2006, 7, 1428–1430 CrossRef CAS PubMed.
- K. D. Alexander and Z. D. Schultz, Anal. Chem., 2012, 84, 7408–7414 CrossRef CAS PubMed.
- T. Deckert-Gaudig, R. Böhme, E. Freier, A. Sebesta, T. Merkendorf, J. Popp, K. Gerwert and V. Deckert, J. Biophotonics, 2012, 5, 582–591 CrossRef PubMed.
- G. Rusciano, G. Zito, G. Pesce and A. Sasso, J. Spectrosc., 2017, 2017, 1–9 CrossRef.
- G. L. Bartolomeo, Y. Zhang, N. Kumar and R. Zenobi, Anal. Chem., 2021, 93, 15358–15364 CrossRef CAS PubMed.
- J. Lombard, Biol. Direct, 2014, 9, 1–35 CrossRef PubMed.
- A. S. Shaw, Nat. Immunol., 2006, 7, 1139–1142 CrossRef CAS PubMed.
- P. Knecht, B. Zhang, J. Reichert, D. A. Duncan, M. Schwarz, F. Haag, P. T. P. Ryan, T.-L. Lee, P. S. Deimel, P. Feulner, F. Allegretti, W. Auwärter, G. Médard, A. P. Seitsonen, J. V. Barth and A. C. Papageorgiou, J. Am. Chem. Soc., 2021, 143, 4433–4439 CrossRef CAS PubMed.
- M. Kim, M. Dygas, Y. I. Sobolev, W. Beker, Q. Zhuang, T. Klucznik, G. Ahumada, J. C. Ahumada and B. A. Grzybowski, J. Am. Chem. Soc., 2021, 143, 1807–1815 CrossRef CAS PubMed.
- M. Kuil, T. Soltner, P. W. N. M. van Leeuwen and J. N. H. Reek, J. Am. Chem. Soc., 2006, 128, 11344–11345 CrossRef CAS PubMed.
- M. L. Merlau, M. del Pilar Mejia, S. T. Nguyen and J. T. Hupp, Angew. Chem., Int. Ed., 2001, 40, 4239–4242 CrossRef CAS PubMed.
- M. C. Feiters, A. E. Rowan and R. J. M. Nolte, Chem. Soc. Rev., 2000, 29, 375–384 RSC.
- K. Kalyanasundaram and M. Gratzel, Coord. Chem. Rev., 1998, 177, 347–414 CrossRef CAS.
- B. Meunier, Chem. Rev., 1992, 92, 1411–1456 CrossRef CAS.
- J.-Y. Gu, Z.-F. Cai, D. Wang and L.-J. Wan, ACS Nano, 2016, 10, 8746–8750 CrossRef CAS PubMed.
- Z. F. Cai, X. Wang, D. Wang and L. J. Wan, ChemElectroChem, 2016, 3, 2048–2051 CrossRef CAS.
- H. Yin, L.-Q. Zheng, W. Fang, Y.-H. Lai, N. Porenta, G. Goubert, H. Zhang, H.-S. Su, B. Ren, J. O. Richardson, J.-F. Li and R. Zenobi, Nat. Catal., 2020, 3, 834–842 CrossRef CAS.
- F. Shao and R. Zenobi, Anal. Bioanal. Chem., 2019, 411, 37–61 CrossRef CAS PubMed.
- X. Wang, S.-C. Huang, T.-X. Huang, H.-S. Su, J.-H. Zhong, Z.-C. Zeng, M.-H. Li and B. Ren, Chem. Soc. Rev., 2017, 46, 4020–4041 RSC.
- J. Lee, K. T. Crampton, N. Tallarida and V. A. Apkarian, Nature, 2019, 568, 78–82 CrossRef CAS PubMed.
- J. Lee, N. Tallarida, X. Chen, L. Jensen and V. A. Apkarian, Sci. Adv., 2018, 4, 1–8 Search PubMed.
- S. Jiang, X. Zhang, Y. Zhang, C. Hu, R. Zhang, Y. Zhang, Y. Liao, Z. J. Smith, Z. Dong and J. G. Hou, Light: Sci. Appl., 2017, 6, e17098–e17098 CrossRef CAS PubMed.
- S. Jiang, Y. Zhang, R. Zhang, C. Hu, M. Liao, Y. Luo, J. Yang, Z. Dong and J. G. Hou, Nat. Nanotechnol., 2015, 10, 865–869 CrossRef CAS PubMed.
- N. Chiang, N. Jiang, D. V. Chulhai, E. A. Pozzi, M. C. Hersam, L. Jensen, T. Seideman and R. P. Van Duyne, Nano Lett., 2015, 15, 4114–4120 CrossRef CAS PubMed.
- R. Zhang, Y. Zhang, Z. C. Dong, S. Jiang, C. Zhang, L. G. Chen, L. Zhang, Y. Liao, J. Aizpurua, Y. Luo, J. L. Yang and J. G. Hou, Nature, 2013, 498, 82–86 CrossRef CAS PubMed.
- N. Chiang, X. Chen, G. Goubert, D. V. Chulhai, X. Chen, E. A. Pozzi, N. Jiang, M. C. Hersam, T. Seideman, L. Jensen and R. P. Van Duyne, Nano Lett., 2016, 16, 7774–7778 CrossRef CAS PubMed.
- K. F. Domke and B. Pettinger, ChemPhysChem, 2009, 10, 1794–1798 CrossRef CAS PubMed.
- Z. F. Cai, L. Q. Zheng, Y. Zhang and R. Zenobi, J. Am. Chem. Soc., 2021, 143, 12380–12386 CrossRef CAS PubMed.
- J. Kučera and A. Gross, Langmuir, 2008, 24, 13985–13992 CrossRef PubMed.
- M. Sun, Z. Zhang, H. Zheng and H. Xu, Sci. Rep., 2012, 2, 2–5 CrossRef PubMed.
- Y. F. Huang, H. P. Zhu, G. K. Liu, D. Y. Wu, B. Ren and Z. Q. Tian, J. Am. Chem. Soc., 2010, 132, 9244–9246 CrossRef CAS PubMed.
- E. M. Van Schrojenstein Lantman, T. Deckert-Gaudig, A. J. G. Mank, V. Deckert and B. M. Weckhuysen, Nat. Nanotechnol., 2012, 7, 583–586 CrossRef CAS PubMed.
- V. Canpean and S. Astilean, Spectrochim. Acta, Part A, 2012, 96, 862–867 CrossRef CAS PubMed.
- J. Ye, J. A. Hutchison, H. Uji-i, J. Hofkens, L. Lagae, G. Maes, G. Borghs and P. Van Dorpe, Nanoscale, 2012, 4, 1606 RSC.
- Z. Zeng, X. Qi, X. Li, L. Zhang, P. Wang and Y. Fang, Appl. Surf. Sci., 2019, 480, 497–504 CrossRef CAS.
- J.-J. Sun, H.-S. Su, H.-L. Yue, S.-C. Huang, T.-X. Huang, S. Hu, M. M. Sartin, J. Cheng and B. Ren, J. Phys. Chem. Lett., 2019, 10, 2306–2312 CrossRef CAS PubMed.
- Y.-F. Huang, M. Zhang, L.-B. Zhao, J.-M. Feng, D.-Y. Wu, B. Ren and Z.-Q. Tian, Angew. Chem., Int. Ed., 2014, 53, 2353–2357 CrossRef CAS PubMed.
- M. Sun, Y. Huang, L. Xia, X. Chen and H. Xu, J. Phys. Chem. C, 2011, 115, 9629–9636 CrossRef CAS.
- N. Hayazawa, T. Yano and S. Kawata, J. Raman Spectrosc., 2012, 43, 1177–1182 CrossRef CAS.
- B. S. Oliveira, B. S. Archanjo, R. Valaski, C. A. Achete, L. G. Cançado, A. Jorio and T. L. Vasconcelos, J. Chem. Phys., 2020, 153, 114201 CrossRef CAS PubMed.
- N. Kumar, S. J. Spencer, D. Imbraguglio, A. M. Rossi, A. J. Wain, B. M. Weckhuysen and D. Roy, Phys. Chem. Chem. Phys., 2016, 18, 13710–13716 RSC.
- L. Opilik, Ü. Dogan, C.-Y. Li, B. Stephanidis, J.-F. Li and R. Zenobi, J. Phys. Chem. C, 2016, 120, 20828–20832 CrossRef CAS.
- T. Umakoshi, K. Kawashima, T. Moriyama, R. Kato and P. Verma, Sci. Rep., 2022, 12, 12776 CrossRef CAS PubMed.
- R. Böhme, M. Mkandawire, U. Krause-Buchholz, P. Rösch, G. Rödel, J. Popp and V. Deckert, Chem. Commun., 2011, 47, 11453 RSC.
| This journal is © The Royal Society of Chemistry 2023 |
