Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synergistic and non-synergistic impact of HAP-based nano fertilizer and PGPR for improved nutrient utilization and metabolite variation in hemp crops†
Agrataben
Vadhel
 a,
Anil
Kumar
a,
Anil
Kumar
 b,
Sabreen
Bashir
b,
Sabreen
Bashir
 a,
Tabarak
Malik
a,
Tabarak
Malik
 *c and
Anand
Mohan
*c and
Anand
Mohan
 *a
*a
aSchool of Bioengineering and Biosciences, Lovely Professional University, Phagwara-144411, Punjab, India. E-mail: anandmohan77@gmail.com
bGene Regulation Laboratory, National Institute of Immunology, New Delhi-110067, India
cDepartment of Biomedical Sciences, Institute of Health, Jimma University, Ethiopia. E-mail: tabarak.malik@ju.edu.et
First published on 11th September 2023
Abstract
Abundant use of nitrogenous fertilizers leads to wasteful expense and environmental degradation through excessive urea availability. We synthesized urea-hydroxyapatite nano fertilizer (UHAPF). Through characterization, we revealed controlled nitrogen release due to reduced hydroxyapatite (HAP) dissolution. Combined application of Bacillus megaterium and Pseudomonas aeruginosa with UHAPF demonstrated improved plant growth in a pot experiment compared to individual treatments, indicating reduced nitrogen loss under field conditions. In a vertical-column setup, gradual nitrogen release in ∼30 days was exhibited by UHAPF. We explored variation in secondary-metabolite formation in industrial cannabis and found that nanofertilizers enhanced nutrient uptake, biofertilizers improved soil health and both affected synergistic cannabinoid production. The presence of 2,4-DTBP during GC-MS prompted additional environmental and health investigations. Our research findings emphasize sustainable approaches for nutrient management, enhanced crop yields and reducing excess nitrogen losses by suggesting novel nano-fertilizer formulations.
Environmental significanceOur research on the synergistic and non-synergistic impact of a HAP-based nano fertilizer and PGPR for improved nutrient utilization and metabolite variation in hemp crops contributes to understanding of nanomaterial interactions with natural systems and their implications for environmental and human health. By developing an environment friendly controlled-release fertilizer, urea hydroxyapatite fertilizer (UHAPF), we addressed the issue of excess urea availability in agricultural fields, which can lead to environmental degradation. We characterized UHAPF and evaluated its urea-release behaviour, offering valuable insights into the controlled release of nano fertilizers and their impact on soil fertility. Furthermore, we investigated the effect of UHAPF combined with selected PGPR on soil fertility, which could prevent further nitrogen leaching into the environment. Our findings contribute to the development of sustainable agricultural practices, enhance understanding of nanomaterial interactions with natural systems and ultimately, safeguarding the environmental and human health. |
Introduction
The global demand for food is escalating due to population growth and shrinking agricultural land. Urbanization and land constraints threaten food security.1 Soil fertility is impacted by factors such as climate change and irrigation. Microorganisms have vital roles in nutrient processes and conventional fertilizers are pivotal for agricultural success.2 Urea and urea-based fertilizers are the preferred nitrogen fertilizers. They have the highest nitrogen percentage and economic value. Conversely, urea utilization has been troublesome due to its low assimilation by plants, which causes severe environmental issues. Nitrate leaching, surface runoff, soil denitrification and volatilization are the main processes contributing to groundwater contamination and global warming because they release nitrous oxide.3 Similarly, phosphorus availability is a concern due to its immobility in soil. Phosphate-solubilizing bacteria can help convert it to a usable form.4In this context, nanotechnology has drawn attention due to its nanoscale size and high surface-to-volume ratio for resolving nutrient losses. To overcome fertilizer-related problems and enhance the efficiency of nitrogen use, nano-synthesized fertilizer has been suggested as an alternative tool and novel solution. Various types of nano-synthesized and slow-release fertilizers have been designed using different nutrient carriers.5–8 However, an environmentally benign fertilizer that does not affect the soil microflora must be developed. To protect urea from rapid release and decomposition, it is complexed into hydroxyapatite (HAP) nanoparticles.9–13 HAP nanoparticles have been exploited as nanocarriers and fertilizers due to their rich surface chemistry with a reactive functional group.14 HAP nanoparticles have been used heavily in biomedical applications because they are nontoxic.15 Hydroxyapatite (Ca10(PO4)6(OH)2) possesses excellent biocompatibility and high biodegradability as well as is a good source of phosphorus.16
Plant growth-promoting rhizobacteria (PGPR) have a vital role in agricultural systems as biofertilizers. PGPR mediate mineralization, nutrient mobilization, denitrification and decomposition. Bacillus megaterium and Pseudomonas aeruginosa have a positive effect on the growth and development of various plant species, such as mustard (Brassica juncea),17 maize (Zea mays), rice (Oryza sativa)18 and tomato (Solanum lycopersicum).19
Cannabis belongs to the Cannabaceae family. It is a notable crop that has been utilized for food, fiber and medicinal purposes for centuries.20,21 It is commonly known as “hemp” or “marijuana”. It is native to Central Asia but has spread to East and South Asia by adapting to various climates.22 Over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 articles have explored its biological aspects23 to reveal over 100 cannabinoids (CBDs), terpenes, flavonoids and essential oils.24 Employed for treating anxiety, sclerosis and chronic pain,25 it is classified into marijuana (1–20%), intermediate (0.3–1.0%) and hemp (0.3%) chemotypes based on delta-9-tetrahydrocannabinol (THC) content.26 The 19th century saw its decline due to psychoactive traits and subsequent prohibition. However, the re-emergence of hemp for textile and biomass is promising. Its cultivation could boost the socioeconomic status of farmers worldwide.27
000 articles have explored its biological aspects23 to reveal over 100 cannabinoids (CBDs), terpenes, flavonoids and essential oils.24 Employed for treating anxiety, sclerosis and chronic pain,25 it is classified into marijuana (1–20%), intermediate (0.3–1.0%) and hemp (0.3%) chemotypes based on delta-9-tetrahydrocannabinol (THC) content.26 The 19th century saw its decline due to psychoactive traits and subsequent prohibition. However, the re-emergence of hemp for textile and biomass is promising. Its cultivation could boost the socioeconomic status of farmers worldwide.27
With phytoattenuation abilities, cannabis detoxifies soil and water by reducing levels of heavy metals, PAHs and organic compounds.28 This adaptable plant thrives along roadways in Indian states such as Punjab, Haryana and Himachal Pradesh. However, research on its industrial and medicinal potentials as a hemp crop and metabolic variability is limited. Its adaptive growth and environmental resilience make it a roadside weed in North India, with environmental factors like humidity, water, salinity and nutrients influencing its accumulation of bioactive compounds.29 In the current study, an environmentally benign urea-HAP fertilizer was synthesized to minimize nutrient runoff. We also investigated the effect of natural and controlled conditions on the phytochemical profile of hemp crops.
Materials and methods
All chemicals and reagents used were of analytical grade. Cetyltrimethylammonium bromide (CTAB), Ca(NO3)2·4H2O, and (NH4)2HPO4 were purchased from Loba Chemie. Microbial culture media were sourced from HiMedia. All other chemicals were obtained from MilliporeSigma. Aqueous solutions were prepared using distilled water.Preparation and characterization of urea hydroxyapatite nano fertilizer (UHAPF)
UHAPF was synthesized according to the method developed by Elhassani30 with modifications. A two-step process was employed to prepare Urea-HAP nanohybrids. Initially, HAP synthesis was done by adding two solutions. The first solution was prepared by dissolving (NH4)2HPO4 (7.92 g) in distilled water followed by the addition of CTAB (1.82 g) and stirring it until a clear solution was formed. Then, the pH was set at 11 with ammonia. Likewise, another solution was prepared with Ca(NO3)2·4H2O (23.615 g) in distilled water. Addition of Ca(NO3)2·4H2O solution in (NH4)2HPO4 under vigorous stirring was done to obtain a milky suspension. This solution was heated for 5 h (80 °C). The precipitate from the solution formed after 24 h was centrifuged followed by water wash. Subsequently, the precipitates were oven-dried at 80 °C to obtain a dry solid powder. In the second step, the prepared HAP powder was sonicated in distilled water until a homogeneous mixture was obtained. Then, the HAP mixture was impregnated with urea solution and the solution was stirred for 15 h. Later, the mixture was dried after being centrifuged and washed with water to remove unreacted urea.The chemical bonding of the synthesized sample was determined using a FTIR spectrometer (Spectrum Two™; Perkin Elmer) with Diamond ATR in the range 600 to 4000 cm−1. X-ray diffraction (XRD) of compounds was done using a diffractometer (D8 Advance XRD; Bruker) equipped with Cu K1 radiation (k = 1.54060 Å) as an X-ray source. The current and voltage of the X-ray tubes were 30 mA and 30 kV, respectively, with a range of 2θ = 10–70°. The morphology and particle size of the sample were revealed by carrying out field emission scanning electron microscopy (FESEM) using a JEOL system. Images were recorded in secondary electron mode. The sample was coated with high-purity gold. Thermal stability of the sample was analysed on TGA 400 apparatus (Perkin Elmer). The sample (∼6 mg) was heated at 10 °C min−1 from room temperature up to 700 °C under airflow. The total amount of nitrogen incorporated with HAP was analysed using the Kjeldahl method.31
Release behaviour
The nitrogen-release behaviour of UHAPF in water was studied in a vertical column setup for 30 days. The tap at the end of the column was covered with cotton. Here, 1 g of urea and UHAPF samples were placed and 50 mL of water was added. At scheduled times, the eluate was collected and centrifuged (500 rpm for 15 min). Then, the supernatant was analysed by a colorimetric method based on para-dimethylaminobenzaldehyde.32 A stock solution containing para-dimethylaminobenzaldehyde solution (20 g L−1) and 4 mL of H2SO4 solution (2 mol L−1) was prepared, from which 2 mL of solution was added to 0.2 mL of urea solution (dm−3). After 15 min, the absorbance was measured at 422 nm using a UV–vis spectrophotometer (LI-2800 Ex; Lasany). The unknown concentration of urea was calculated using a standard curve. Different kinetic models were used to understand how urea was released over time.PGPR strain: phosphate solubilization potential and inocula preparation
B. megaterium MTCC1684 and P. aeruginosa MTCC7453 strains were tested for solubilization of tricalcium phosphate. These bacterial strains were obtained from Microbial Type Culture Collection and Gene Bank from Chandigarh (India). Bacterial strains were re-cultured on nutrient agar medium containing water (1 L) as well as (in g L−1) peptone (5), HM peptone B# (1.5), yeast extract (1.5), sodium chloride (5) and agar (15) at pH 7.4 ± 0.2. NBRIP medium was used for screening the capacity for solubilizing bacterial strains from insoluble phosphate.33 Tricalcium phosphate was used as an insoluble phosphorus source. The NBRIP medium contained (in g L−1) glucose (10), Ca3(PO4)2 (5), KCL (1), NH4SO4, (0.5), MgSO4 7H2O (0.1), MnSO4 (0.01) yeast extract (0.5), bromocresol purple (0.1) and agar (20). Bromocresol purple is used as a pH indicator in microbial growth media. This indicator was yellow below pH 3.8 and purple above pH 5.4 (additional file 1: Fig. S1†). For further experimental purposes, bacterial strains inocula were harvested from sterilized nutrient broth culture medium which contained (in g L−1) peptone (5), HM peptone B# (1.5), yeast extract (1.5) and sodium chloride (5) at pH 7.4 ± 0.2. The culture (108 CFU mL−1) was harvested (30 °C for 48 h) during the log phase at 120 rpm on a shaker. Peat was used as a carrier for longer viability and high effectiveness of microbial culture.34 Assessment of the viability of B. megaterium and P. aeruginosa in peat samples after 48 h of incubation confirmed their establishment in soil.Plant material and growth condition
To observe the synergistic effect of the PGPR strain with the synthesized UHAP fertilizer, a pot-culture experiment was carried out by comparing the growth and yield of Cannabis sativa L. The plant was collected from the roadside and its identification was done by Council Of Scientific And Industrial Research-Indian Institute Of Integrative Medicine (CSIR-IIIM) Jammu (accession number = 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 832). The herbarium was submitted to a national referral facility: Janaki Ammal Herbarium CSIR-IIIM. The study was conducted at Lovely Professional University (Punjab, India) under controlled conditions at a mean temperature of 24–28 °C. Soil analyses were done at the Soil Testing Laboratory of Punjab Agriculture University (Ludhiana, India). Soil analyses estimated the pH at 8.1 with mineral contents in their available form (in kg acre−1) P (7.8 kg), potash (92 kg), Zn (0.83), Fe (7.51), Mg (6.71) and Cu (0.39). Pots were filled with 5 kg of unsterilized field soil. The two PGPR strains were used alone and/or in combination with UHAPF. The trial was conducted as a complete randomized design with 21 pots including seven treatments: (1) control, (2) urea (UF); (3) synthesized UHAPF; (4) B. megaterium (B1); (5) P. aeruginosa (B2); (6) B1+UHAPF; (7) B2+UHAPF. The treatment was given in the vegetative stage after 6 weeks. The amount of fertilizer given to the plant was calculated according to the weight of the soil.
832). The herbarium was submitted to a national referral facility: Janaki Ammal Herbarium CSIR-IIIM. The study was conducted at Lovely Professional University (Punjab, India) under controlled conditions at a mean temperature of 24–28 °C. Soil analyses were done at the Soil Testing Laboratory of Punjab Agriculture University (Ludhiana, India). Soil analyses estimated the pH at 8.1 with mineral contents in their available form (in kg acre−1) P (7.8 kg), potash (92 kg), Zn (0.83), Fe (7.51), Mg (6.71) and Cu (0.39). Pots were filled with 5 kg of unsterilized field soil. The two PGPR strains were used alone and/or in combination with UHAPF. The trial was conducted as a complete randomized design with 21 pots including seven treatments: (1) control, (2) urea (UF); (3) synthesized UHAPF; (4) B. megaterium (B1); (5) P. aeruginosa (B2); (6) B1+UHAPF; (7) B2+UHAPF. The treatment was given in the vegetative stage after 6 weeks. The amount of fertilizer given to the plant was calculated according to the weight of the soil.
Soil and yield analyses
After treatment, samples were taken after 30 days to estimate growth and biochemical activities, as shown below.Estimation of photosynthetic pigments, total protein, available nitrogen and available phosphorus content
Photosynthetic pigments such as carotenoid and chlorophyll were estimated by the methods described by Lichtenthaler35 and Arnon,36 respectively. Eighty percent of an acetone extract of the plant sample was centrifuged to measure absorbance at 663 nm and 645 nm for chlorophyll, and 480 nm and 510 nm for carotenoids using a UV-vis spectrophotometer (LI-2800 Ex). We used several equations for calculations: chlorophyll a (mg g−1 FW) = (Abs663 × 12.7) − (Abs645 × 2.69) × V/(1000 × W); chlorophyll b (mg g−1 FW) = (Abs645 × 22.9) − (Abs663 × 4.68) × V/(1000 × W); total chlorophyll content (mg g−1 FW) = (Abs645 × 22.2) + (Abs663 × 8.03) × V/(1000 × W); total carotenoid content (mg g−1 FW) = 7.6 (Abs480) − 1.49 (Abs510) × (V/d × W × 1000). The protein content of cannabis leaves was estimated using the Lowry method.37 First, 0.1 g of a fresh leaf sample was homogenized with phosphate-buffered saline using an ice-cold mortar pestle and then the sample was centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 20 min, 4 °C). The supernatant was used to measure protein content by the Lowry method using bovine serum albumin (BSA) as a standard. The available nitrogen of soil after treatment was determined by the alkaline permanganate method.38 The available phosphorus was determined by the method described by Olsen.39
000 rpm, 20 min, 4 °C). The supernatant was used to measure protein content by the Lowry method using bovine serum albumin (BSA) as a standard. The available nitrogen of soil after treatment was determined by the alkaline permanganate method.38 The available phosphorus was determined by the method described by Olsen.39
Extraction and analyses of the volatile compounds in hemp
The traditional maceration extraction procedure was applied to analyse the bioactive compounds present in hemp leaves.40 Dried powder of hemp leaves (1 g) was extracted with 100 mL of methanol at room temperature for 24 h. Then, the solution was paper-filtered and evaporated to dryness. The obtained extractive compound was stored in an airtight bottle at 4 °C. The analysis was undertaken on a gas chromatograph (GCMS-TQ8040 NX; Shimadzu) with a mass spectra detector. The compound was separated on SH-RXi-5Sil MS crosslinked to 5% diphenyl/95% dimethyl polysiloxane (30 m × 0.32 mm, 0.25 μm) capillary column. The initial column temperature was set at 40 °C, then increased to 250 °C at the rate of 7 °C min−1, which was held for 3 min. Helium was used as the carrier gas (flow rate = 1 mL min−1). The detection and identification of compounds in the extract was achieved using the National Institute of Standards and Technology spectral database. Detection was done in Q3 scan acquisition mode within an m/z range of 40 to 500.41 The proportion of an individual component is presented as a relative peak area percentage of the total peak percentage.Statistical analyses
Statistical comparisons were conducted using the SPSS 22 (IBM). Data were subjected to a one-way analysis of variance (ANOVA) and Duncan's multiple comparison test. P < 0.05 was considered significant. The longevity of N release was calculated by regression equations and coefficients. The molecular structure was visualized using ChemAxon (MarvinSketch).Results and discussion
Characterization of nanofertilizers with various analytical instruments
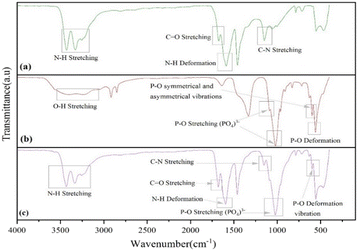
The FTIR spectra of UF and UHAPF are depicted in Fig. 1. The O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–NH2 functional group, which is made up of the N–H group, C
C–NH2 functional group, which is made up of the N–H group, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and –CN bonds, had an absorption peak for urea fertilizer at 3428.82 cm−1, 1673.66 cm−1 and 1148.75 cm−1, respectively. The stretching and deformation vibrations of the N–H bond in pure urea were observed at absorption bands of 3428.82 cm−1 and 1588.20 cm−1. Through possible band shifting, the potential interfacial interaction between urea and HAP was monitored. Stretching of the phosphate group (PO43−) was responsible for the sharp and intense absorption bands at 1088.00 cm−1 and 1020.73 cm−1. The peak at 598.59 cm−1 was typical of P–O deformation vibrations, whereas the signal at 962.38 cm−1 was caused by P–O symmetrical and asymmetrical stretching vibrations. The band observed at 3403.20 cm−1 was characteristic of the stretching vibration of hydroxyl groups of the HAP structure. The absorption bands observed at 3428.82 cm−1 and 1588.20 cm−1, on stretching and deformation vibrations of N–H bonds in pure urea, were shifted to higher frequencies of 3431.64 cm−1 and 1595.14 cm−1 in the spectrum of urea-coated HAP. These data suggested the existence of strong hydrogen bonding between the O–H groups of HAP and N–H groups of urea.42,43
O and –CN bonds, had an absorption peak for urea fertilizer at 3428.82 cm−1, 1673.66 cm−1 and 1148.75 cm−1, respectively. The stretching and deformation vibrations of the N–H bond in pure urea were observed at absorption bands of 3428.82 cm−1 and 1588.20 cm−1. Through possible band shifting, the potential interfacial interaction between urea and HAP was monitored. Stretching of the phosphate group (PO43−) was responsible for the sharp and intense absorption bands at 1088.00 cm−1 and 1020.73 cm−1. The peak at 598.59 cm−1 was typical of P–O deformation vibrations, whereas the signal at 962.38 cm−1 was caused by P–O symmetrical and asymmetrical stretching vibrations. The band observed at 3403.20 cm−1 was characteristic of the stretching vibration of hydroxyl groups of the HAP structure. The absorption bands observed at 3428.82 cm−1 and 1588.20 cm−1, on stretching and deformation vibrations of N–H bonds in pure urea, were shifted to higher frequencies of 3431.64 cm−1 and 1595.14 cm−1 in the spectrum of urea-coated HAP. These data suggested the existence of strong hydrogen bonding between the O–H groups of HAP and N–H groups of urea.42,43
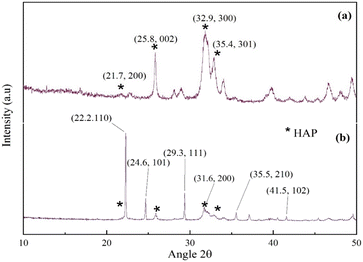
The powder XRD patterns of synthesized HAP and UHAPF are depicted in Fig. 2. The XRD pattern of HAP, with a well-crystalline hexagonal structure, followed the pattern observed in PDF card number 01-076-694 from JCPDS (monoclinic, a = 9.4214 Å, b = 18.8428 Å, c = 6.8814 Å). According to Bramhe,44 the synthesis of HAP via the solution combustion method by the addition of ammonia solution can be explored for coating application material. Urea peaks coincided with pure urea without peak shifting, indicating that urea had been immobilized on the HAP support without disrupting the crystal structure of urea.45 The XRD pattern of urea peaks is indexed as 2θ = 22.2, 24.6, 29.3, 35.5, 41.5 and 31.6, which was attributed to the (110), (101), (111), (210), (102) and (200) crystal planes, respectively, reported in JCPDS file number 00-031-1979. The peak at 2θ = 22.2° had the highest intensity and was the most prominent peak of the (110) plane of the tetragonal phase matching with JCPDS file number 00-031-1979. Moreover, neither before nor after impregnation with urea was a significant shift observed in HAP peak positions in UHAPF synthesis.
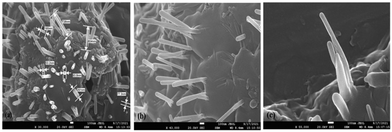
SEM revealed the morphology of UHAPF (Fig. 3). The nano form of HAP exhibited a rod-like morphology and hexagonal crystal structure as reported in the literature.10 Aghayan and Rodríguez (2012) investigated the morphologic changes of HA nanoparticles according to the urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HNO3 ratio. They stated that rod-shaped nanoparticles developed when the urea
HNO3 ratio. They stated that rod-shaped nanoparticles developed when the urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HNO3 was close to 1. This phenomenon resulted from the complete oxidation of urea, which releases more energy in the process.46 The observed morphology, therefore, confirmed the formation of nanohybrids between urea and HAP.
HNO3 was close to 1. This phenomenon resulted from the complete oxidation of urea, which releases more energy in the process.46 The observed morphology, therefore, confirmed the formation of nanohybrids between urea and HAP.
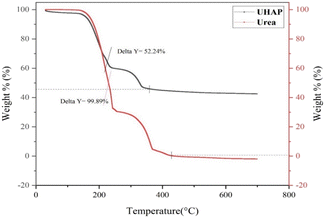
The thermogravimetric curve of synthesized UHAPF (Fig. 4) revealed the thermal decomposition of urea and synthesized UHAPF. Urea is a thermally unstable compound.47 The thermal decomposition of urea started before the melting point (132.5 °C) until its complete oxidation at 400 °C. The sample was heated at a constant heating rate until 700 °C and held for 1.0 min at 700 °C. UHAPF degradation of ∼52 wt% remained at this temperature, indicating that 48 wt% of urea was incorporated into HAP. Urea decomposition increased when it was supported by HAP, indicating that HAP may serve as a support but also as a catalyst for the thermal degradation of urea.
Estimation of release behaviour
Total nitrogen was calculated using the Kjeldahl method. The total nitrogen content of pure urea is 32.72%. Upon analysis, it was found that UHAPF contained 13.54% nitrogen. Hence, nearly about half of urea was incorporated in HAP. Estimation of nitrogen leaching over 30 days provided a preliminary indication of a novel controlled-release formulation of synthesized UHAPF compared with pure urea. The nitrogen release rate of UHAPF was strongly dependent upon the coating material and formulation. UHAPF in an aqueous medium showed a much slower release rate as compared with that of pure urea (Fig. 5). The moderately strong bond of HAP and urea formed through its amine and carbonyl groups confirmed that UHAPF met the nitrogen demand of the plant.48 The release profile of UHAPF was analyzed using kinetic-release models based on the best fit. In the Korsmeyer–Peppas model, the constant value “kKP” for UF and UHAPF was 78.7 and 3.2, respectively, and represented the release rate constant. The R2 for UF and UHAPF was 0.99 and 0.97, respectively. The MSC for UF was 2.67, and for UHAPF it was 3.01. The AIC for UF was 37.72, and for UHAPF it was 48.2. The value of “n” for the Korsmeyer–Peppas model was 0.05 for UF and 0.88 for UHAPF. The Korsmeyer–Peppas model appeared to be the best-fit model for UF and UHAPF systems because it had the highest R2 values. This model assumed that urea release was controlled by diffusion and polymer relaxation. UF had a higher R2 than UHAPF and the lowest AIC, indicating it was the best fit for this model. The n-value for UHAPF was close to 1, indicating Fickian diffusion. The n-value for UF was much smaller, indicating anomalous diffusion.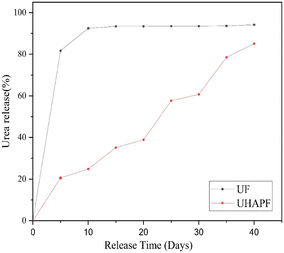 | ||
| Fig. 5 The release rate of urea fertilizer (UF) and UHAP fertilizer (UHAPF) as a function of time at 28°C. | ||
The high R2 values indicated that the model explained a noteworthy proportion of the variation in drug-release data. The low mean square errors and AIC values further supported the suitability of this model. The exponent (n) values also suggested that the drug-release mechanism was non-Fickian or anomalous in both fertilizers. These data indicated the similarity with the water-soluble drug molecules from a homogeneous matrix. In slow-release urea formulations, the release is controlled by diffusion. Similarly, Xiaoyu et al. enabled controlled release of urea by adding an organic polymer and bentonite to create a structure around urea that was released slowly. Double-exponent equations and the Peppas model were used to analyze release behaviour: dissolution and erosion of the lattice structure surrounding urea molecules were the main factors.49
Changes in growth and yield parameters
Effect on photosynthetic pigments and protein content of C. sativa after treatment
The content of photosynthetic pigments in the leaves of C. sativa L. (chlorophyll a, chlorophyll b, total chlorophyll and carotenoids) was increased significantly in treated plants as compared with the control. High chlorophyll and carotenoid contents of 3.39 mg g−1 FW and 3.10 mg g−1 FW, respectively, in combined exposure of synthesized fertilizer and B. megaterium as compared with sole application (control treatment) was noted (Fig. 6A). Due to the slow release of nitrogen in soil, plant uptake of nitrogen was greater from UHAPF than normal urea fertilizer, which suggested that a lower amount of fertilizer leached into water. In addition, B1 and B2 showed higher yields than control because PGPR solubilizes insoluble phosphorus and fixes the nitrogen present in the soil to benefit the plant. Co-inoculation of PGPR with UHAPF was more effective and produced a higher yield than their sole application. Likewise, the content of protein was also increased on giving the same treatment to the plant. This growth improvement was associated with the capacity for phosphate solubilization and production of organic acids (Fig. 6B). B. megaterium and P. aeruginosa act as bio-fertilizers for the growth and development of plants. Kang et al. studied the use of B. megaterium for increasing the growth of mustard plants. B. megaterium was found to increase the root length, shoot length and fresh weight of B. megaterium. This bacterial interaction changed the biochemical pathways in plants by increasing the contents of photosynthetic pigments and protein.17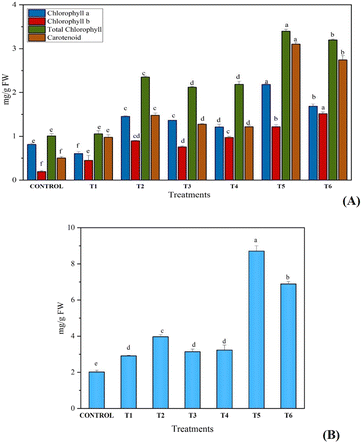 | ||
| Fig. 6 Effect on A) photosynthetic pigments and B) protein content of Cannabis sativa L. after sole and combine treatment of synthesized fertilizer and biofertilizer. | ||
The solubilization of HAP in soil is affected by a “microbial consortia” of phosphate-solubilizing microorganisms (PSMs) which also enhance total phosphorus absorption and increase root proliferation, plant biomass and other isolates significantly.52 Sedri et al. undertook comparative analysis of PGPR and chemical fertilizers for rain-fed wheat. They suggested that PGPR could be a significant alternative to chemical fertilizers and help to improve the production of cereals in cool, rain-fed cultivation systems.53 PGPR solubilized insoluble forms of phosphorus in soil but also improved soil fertility after the plant had been removed from soil (Table 1).
| Treatment | Control | T1 | T2 | T3 | T4 | T5 | T6 |
|---|---|---|---|---|---|---|---|
| UF | UHAPF | B1 | B2 | UHAPF+B1 | UHAPF+B2 | ||
| Data are presented as mean ± S.E. (n = 3). At the P < 0.05 level, different letters on each error bar are statistically significant. Treatments: 1) control, 2) T1 – Urea, 3) T2 – Synthesized Urea Hydroxyapatite Nanofertilizer (UHAPF), 4) T3 – Bacillus megaterium (B1), 5) T4 – Pseudomonas aeruginosa (B2), 6) T5 – B1+UHAPF, 7) T6 – B2+UHAPF. | |||||||
| Available nitrogen (kg ha−1) | 245.65 ± 13.066f | 284.85 ± 13.66d,e | 297.92 ± 0d | 363.25 ± 13.066c | 324.05 ± 13.06c,d | 454.72 ± 22.63b | 546.18 ± 13.066a |
| Available phosphorus (kg ha−1) | 6.56 ± 0.245e | 6.535 ± 0.155e | 8.68 ± 0.097d | 11.75 ± 0.5914c | 12.13 ± 1.1303c | 28.72 ± 0.1297a | 26.90 ± 0.5205b |
Bioactive compounds in natural vs. controlled conditions
The presence of bioactive compounds in cannabis was evaluated using gas chromatography-mass spectrometry (GC-MS). This study revealed that CBDs, saturated and unsaturated fatty acids and 2,4-di-tert-butylphenol (2,4-DTBP) were present in the natural conditions. In controlled conditions, various types of CBDs, including THC and fatty acids, were identified (Fig. 7). The relative percent area of 2,4-DTBP was significantly higher (25.61%) compared with that of other compounds in the natural conditions. However, in a pot experiment carried out under the controlled conditions, a negligible amount (0.20%) of 2,4-DTBP was noted. It is an allelochemical and has not been reported previously in the context of hemp research. Allelochemicals suppress the growth of neighboring plants and provide a competitive advantage to the producing species. Several studies have shown that 2,4-DTBP occurs naturally in medicinal plants and exhibits potent herbicidal properties by altering the chloroplast ultrastructure of weedy plants and reducing the physiological activity of other plants.54 It also possesses a potential pre-emergent herbicidal activity by acting as a strong root inhibitor.55 A recent study has suggested that 2,4-di-tert-butylphenol detected in the culm and leaf extracts of Pennisetum purpureum can prevent the root growth of Hedyotis verticillata (a common broadleaf weed in oil-palm plantations) and Leptochloa chinensis (a grassy weed often found in rice fields).56 However, allelopathy is a complex phenomenon which involves various allelochemicals with different modes of action. It is possible that 2,4-DTBP is one of many allelochemicals present in plants that may have a role in promoting the survival, growth and development of plant species under natural conditions.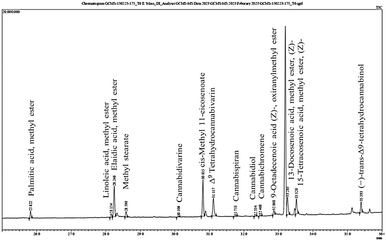 | ||
| Fig. 7 The secondary metabolite composition under controlled condition graph of Cannabis sativa L. extract used in this study. | ||
The secondary-metabolite content of C. sativa L. extracted by maceration was analysed by GC-MS and identified from an MS spectral library (additional file: Table S1†). The identified compounds were divided into three categories based on their chemical nature: fatty acid methyl esters, cannabis-specific compounds, and others. CBD compounds are not classified as alkaloids due to their lack of nitrogen atom in its structure and are considered instead as terpenophenolic compounds. Some identified compounds are specific to the cannabis plant, such as cannabidiol, Δ9tetrahydrocannabivarin (THCV), cannabichromene (CBC), cannabispiran (CBS), and dronabinol. Fatty acid methyl esters such as palmitic acid, linoleic acid, methyl elaidate, methyl stearate, cis-methyl 11-eicosenoate, methyl erucate, glycidyl oleate and methyl nervonate were also detected in C. sativa L.
Semi-quantitative data for bioactive compounds from C. sativa L.
The highest amounts of CBD (13.27%), THCV (14.33%), and THC (19.34%) were detected in the treatment group, which was a combination of P. aeruginosa and UHAPF (additional file: Table S2†). CBS, CBC and cannabidivarine (CBDV) were not detected in the control group. The application of bacterial inoculants (T3 and T4) also changed the chemical profile of the cannabis plant. The most significant increases in T3 were observed for CBS, CBC and CBDV. The most significant increases in T4 were observed for 15-tetracosenoic acid, methyl ester, (Z)-, 13-docosenoic acid, methyl ester, (Z)- and THC. The increase in percent relative peak area of these compounds was due to the positive impact of P. aeruginosa and B. megaterium on soil fertility and plant growth. Schott et al. revealed that hemp contains various microbial endophytes, such as Pseudomonas species, Pantoea species and Bacillus species, with the ability to produce siderophores, cellulase and aid phosphate solubilization. Pseudomonas species are the most widespread and best candidates for producing bioactive compounds in hemp.57 In addition, the application of synthesized urea fertilizer (T2, T4, T5) led to a significant increase in percent relative peak area compared with that for control in some compounds.The concentrations of CBD, THC and THCV increased from individual treatments to combined treatments, whereas the concentrations of CBC and CBDV showed inconsistent changes. Other compounds, such as methyl stearate, cis-methyl 11-eicosenoate and glycidyl oleate were also found in samples. The concentration of these compounds showed varying trends with increasing treatments. The increase in concentrations of CBD, THC and THCV in combined treatments could have been due to activation of CBD biosynthesis pathways. CBD biosynthesis in hemp plants is influenced by genetics, environmental conditions and cultivation practices. Factors that can influence CBD content include temperature, humidity and nutrient availability. On the other hand, the decrease in fatty acid methyl esters could indicate a reduction in lipid biosynthesis, possibly as an adaptive response to these treatments. Overall, the results suggested that the conditions used in T3 and T5 could promote the synthesis of beneficial compounds in the cannabis plant. The application of different treatments to cannabis plants can affect their secondary-metabolite composition. Cockson et al. explored the impacts of different concentrations of phosphorus on the growth, development and quality of C. sativa and monitored plant height, leaf-tissue mineral-nutrient concentrations, diameter and the final weight of fresh flower buds. They also examined the CBD and terpene levels in flowers to assess the impact of phosphorus fertility on floral quality. They suggested that phosphorus concentrations had a substantial effect on the growth and development of cannabis plants.58 The increase in the percentage of certain compounds, such as CBDs and fatty acid esters, may have implications on the medicinal and industrial use of cannabis plants. Also, using bacterial inoculants and synthesized fertilizers may be a viable method for increasing the production of specific compounds in cannabis plants.
Atoloye et al. studied different hemp varieties to examine the effect of nitrogen fertilizer on CBD yield and bud biomass in field conditions. Their findings suggested that CBD content was influenced by nitrogen fertilizer.59 Likewise, Caplan et al. tested four concentrations of organic fertilizer: the highest CBD content was observed at an organic fertilizer rate of 389 mg N/L.60 Hemp is an annual herbaceous plant that demands high nitrogen supply for increasing plant yield and regulates terpenoid and CBD profiles.50
The detection of psychoactive and non-psychoactive compounds and biomass can vary depending on the cannabis strain, extraction method and solvent selection. The composition and quantity of secondary metabolites of cannabis plants mainly depends on environmental conditions, genetic makeup and cultivation conditions.61 However, there is a limited understanding of how environmental and cultivation conditions affect the regulation of secondary metabolism in the plant.50,61 Cannabidiol, cannabidivarine and CBC are non-psychoactive CBDs with potential anti-inflammatory, analgesic, anxiolytic and anti-epileptic effects, and are found in relatively low concentrations in the hemp plant. Out of all CBDs, cannabidiol is most abundant in hemp plants and is often extracted and used in the production of cannabis-based medications and supplements. Dronabinol is a synthetic form of THC, the main psychoactive compound in drug-type or fibre-type cannabis plants. The US Food and Drug Administration approves its usage in the treatment of chemotherapy-induced nausea and vomiting, as well as for stimulating appetite in patients with AIDS-related wasting syndrome. Dronabinol is not present naturally in hemp plants. However, its development and approval for medical use highlight the potential benefits of CBDs and the importance of continued research in this field.62
Conclusions
The scope of conventional fertilizers has become limited due to the low efficiency of crop nutrients and heavy loss into the environment. We aimed to synthesize UHAPF to improve the efficiency of nitrogen use and crop productivity. The characterization techniques confirmed the successful synthesis of UHAPF. A lower amount of dissolution of HAP in aqueous media compared with chemical fertilizer provided greater nitrogen efficiency through controlled release as well as is a novel phosphorus source. With half of the demand for pure urea, the synthesized UHAPF increased the plant yield. A combination of B. megaterium and P. aeruginosa with UHAPF was the best treatment for the plant. Our findings indicate that joint application of PGPR and UHAPF was more effective than single inoculation treatment, and could help to reduce excess nitrogen loss under field conditions.Hemp is a multipurpose and valuable crop for industrial applications such as in the production of fibre, seeds and medicinal compounds. As the popularity of cannabis continues to rise, it becomes crucial to be aware about its established benefits and potential drawbacks. Therefore, extensive research is necessary to investigate the therapeutic properties of its active components, such as THC and CBDs. Nanofertilizer and biofertilizers have potential impact on the CBD content of cannabis plants. Nanofertilizers provide more efficient uptake and utilization of nutrients. Biofertilizers can improve soil health and nutrient availability. Both types of fertilizers affect the production of CBDs, which are synthesized by the plant in response to environmental cues and nutrient availability. The detection of 2,4-DTBP in this specie in natural conditions highlights the need for further investigation into the potential implication of its presence and potential toxicity to the environment and human health.
Author contributions
Conception: Anand Mohan and Tabarak Malik. Design of the work: Agrataben Vadhel and Anil Kumar. Manuscript drafting: Agrataben Vadhel and Sabreen Bashir.Conflicts of interest
All authors declare no conflict of interest.Acknowledgements
The authors are grateful to Lovely Professional University for providing facilities.References
- T. Zhou, Y. Wang, S. Huang and Y. Zhao, Sci. Total Environ., 2018, 615, 422–430 CrossRef CAS PubMed.
- H. Guo, J. C. White, Z. Wang and B. Xing, Curr. Opin. Environ. Sci. Health, 2018, 6, 77–83 CrossRef.
- C. P. Witte, Plant Sci., 2011, 180, 431–438 CrossRef CAS PubMed.
- M. Everaert, R. Warrinnier, S. Baken, J. P. Gustafsson, D. De Vos and E. Smolders, ACS Sustainable Chem. Eng., 2016, 4, 4280–4287 CrossRef CAS.
- G. D. Feng, Y. Ma, M. Zhang, P. Y. Jia, L. H. Hu, C. G. Liu and Y. H. Zhou, Prog. Org. Coat., 2019, 133, 267–275 CrossRef CAS.
- A. S. Giroto, G. G. Guimarães, L. A. Colnago, A. Klamczynski, G. Glenn and C. Ribeiro, J. Cleaner Prod., 2019, 217, 448–455 CrossRef CAS.
- A. Rashidzadeh and A. Olad, Carbohydr. Polym., 2014, 114, 269–278 CrossRef CAS PubMed.
- X. Yu and B. Li, Particuology, 2019, 45, 124–130 CrossRef CAS.
- P. Fincheira, N. Hoffmann, G. Tortella, A. Ruiz, P. Cornejo, M. C. Diez, A. B. Seabra, A. Benavides-Mendoza and O. Rubilar, Nanomaterials, 2023, 13, 1978 CrossRef CAS PubMed.
- N. Kottegoda, C. Sandaruwan, G. Priyadarshana and A. Siriwardhana, ACS Nano, 2017, 11(2), 1214–1221 CrossRef CAS PubMed.
- N. L. Fernando, D. T. N. Rathnayake, N. Kottegoda, J. K. D. S. Jayanetti, V. Karunaratne and D. R. Jayasundara, Langmuir, 2021, 37, 6691–6701 CrossRef CAS PubMed.
- M. R. Maghsoodi, N. Najafi, A. Reyhanitabar and S. Oustan, Geoderma, 2020, 379, 114644 CrossRef CAS.
- M. Noruzi, P. Hadian, L. Soleimanpour, L. Ma'mani and K. Shahbazi, Chem. Biol. Technol. Agric., 2023, 10, 71 CrossRef CAS.
- N. Madusanka, C. Sandaruwan, N. Kottegoda, D. Sirisena, I. Munaweera, A. De Alwis, V. Karunaratne and G. A. J. Amaratunga, Appl. Clay Sci., 2017, 150, 303–308 CrossRef CAS.
- W. P. S. L. Wijesinghe, M. M. M. G. P. G. Mantilaka, R. M. G. Rajapakse, H. M. T. G. A. Pitawala, T. N. Premachandra, H. M. T. U. Herath, R. P. V. J. Rajapakse and K. G. U. Wijayantha, RSC Adv., 2017, 7, 24806–24812 RSC.
- L. Xiong, P. Wang and P. M. Kopittke, Geoderma, 2018, 323, 116–125 CrossRef CAS.
- S. M. Kang, R. Radhakrishnan, Y. H. You, G. J. Joo, I. J. Lee, K. E. Lee and J. H. Kim, Indian J. Microbiol., 2014, 54, 427–433 CrossRef CAS PubMed.
- X. Liu, H. Zhao and S. Chen, Curr. Microbiol., 2006, 52, 186–190 CrossRef CAS PubMed.
- A. O. Adesemoye and E. O. Ugoji, Arch. Phytopathol. Plant Prot., 2009, 42, 188–200 CrossRef CAS.
- E. M. J. Salentijn, Q. Zhang, S. Amaducci, M. Yang and L. M. Trindade, Ind. Crops Prod., 2015, 68, 32–41 CrossRef.
- N. Bernstein, J. Gorelick, R. Zerahia and S. Koch, Front. Plant Sci., 2019, 10, 736 CrossRef PubMed.
- R. Clarke and M. Merlin, Cannabis: evolution and ethnobotany, Univ of California Press, 2016 Search PubMed.
- A. Hazekamp, J. T. Fischedick, M. L. Díez, A. Lubbe and R. L. Ruhaak, in Comprehensive Natural Products II, Elsevier, 2010, pp. 1033–1084 Search PubMed.
- F. Pellati, V. Brighenti, J. Sperlea, L. Marchetti, D. Bertelli and S. Benvenuti, Molecules, 2018, 23(10), 2639 CrossRef PubMed.
- J. Gonçalves, T. Rosado, S. Soares, A. Simão, D. Caramelo, Â. Luís, N. Fernández, M. Barroso, E. Gallardo and A. Duarte, Medicines, 2019, 6, 31 CrossRef PubMed.
- S. A. Ahmed, S. A. Ross, D. Slade, M. M. Radwan, F. Zulfiqar and M. A. ElSohly, J. Nat. Prod., 2008, 71, 536–542 CrossRef CAS PubMed.
- M. K. Nath, Chemistry Proceedings, 2022, 10(1), 14 Search PubMed.
- N. B. Rioba, F. M. Itulya, M. Saidi, N. Dudai and N. Bernstein, J. Appl. Res. Med. Aromat. Plants, 2015, 2, 21–29 Search PubMed.
- D. S. Seigler, Plant Secondary Metabolism, Springer US, Boston, MA, 1998, vol. 21 Search PubMed.
- C. E. Elhassani, Y. Essamlali, M. Aqlil, A. M. Nzenguet, I. Ganetri and M. Zahouily, Environ. Technol. Innovation, 2019, 15, 100403 CrossRef.
- J. Kjeldahl, Z. Anal. Chem., 1883, 22(1), 366–382 CrossRef.
- M. T. Knorst, R. Neubert and W. Wohlrab, J. Pharm. Biomed. Anal., 1997, 15, 1627–1632 CrossRef CAS PubMed.
- R. Gupta, R. Singal, A. Shankar, R. C. Kuhad and R. K. Saxena, J. Gen. Appl. Microbiol., 1994, 40, 255–260 CrossRef CAS.
- A. Novinscak and M. Filion, Front. Sustain. Food Syst., 2020, 4, 1–10 CrossRef.
- H. K. Lichtenthaler and A. R. Wellburn, Biochem. Soc. Trans., 1983, 11, 591–592 CrossRef CAS.
- D. I. Arnon, Plant Physiol., 1949, 24, 1–15 CrossRef CAS PubMed.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and R. J. Randall, J. Biol. Chem., 1951, 193, 265–275 CrossRef CAS PubMed.
- V. V. Subbaiah and G. K. Asija, Curr. Sci., 1956, 26, 258–260 Search PubMed.
- S. R. Olsen, Estimation of available phosphorus in soils by extraction with sodium bicarbonate, US Department of Agriculture, 1954 Search PubMed.
- J. Pande and S. Chanda, J. Pharm. Biomed. Anal., 2020, 186, 113347 CrossRef CAS PubMed.
- A. Leghissa, Z. L. Hildenbrand, F. W. Foss and K. A. Schug, J. Sep. Sci., 2018, 41, 459–468 CrossRef CAS PubMed.
- B. Roshanravan, S. M. Soltani, F. Mahdavi, S. A. Rashid and M. K. Yusop, Chem. Speciation Bioavailability, 2014, 26, 249–256 CrossRef.
- N. Madusanka, C. Sandaruwan, N. Kottegoda, D. Sirisena, I. Munaweera, A. De Alwis, V. Karunaratne and G. A. J. Amaratunga, Appl. Clay Sci., 2017, 150, 303–308 CrossRef CAS.
- S. N. Bramhe, H. C. Lee, M. C. Chu, J. K. Ryu, A. Balakrishnan and T. N. Kim, Han'guk Chaelyo Hakhoechi, 2015, 25, 492–496 CAS.
- S. Bakshi, C. Banik, D. A. Laird, R. Smith and R. C. Brown, ACS Sustainable Chem. Eng., 2021, 9, 8222–8231 CrossRef CAS.
- M. A. Aghayan and M. A. Rodríguez, Mater. Sci. Eng., C, 2012, 32, 2464–2468 CrossRef CAS.
- S. Chen, M. Yang, C. Ba, S. Yu, Y. Jiang, H. Zou and Y. Zhang, Sci. Total Environ., 2018, 615, 431–437 CrossRef CAS PubMed.
- G. P. Gunaratne, N. Kottegoda, N. Madusanka, I. Munaweera, C. Sandaruwan, W. M. G. I. Priyadarshana, A. Siriwardhana, B. A. D. Madhushanka, U. A. Rathnayake and V. Karunaratne, Indian J. Agric. Sci., 2016, 86, 494–499 CAS.
- N. Xiaoyu, W. Yuejin, W. Zhengyan, W. Lin, Q. Guannan and Y. Lixiang, Biosyst. Eng., 2013, 115, 274–282 CrossRef.
- A. Saloner and N. Bernstein, Front. Plant Sci., 2020, 11, 572293 CrossRef PubMed.
- S. Landi, R. Berni, G. Capasso, J. F. Hausman, G. Guerriero and S. Esposito, Int. J. Mol. Sci., 2019, 20, 1–16 Search PubMed.
- M. Amri, D. Mateus, M. Gatrouni, M. R. Rjeibi, N. Asses and C. Abbes, Appl. Biosci., 2022, 1, 179–197 CrossRef.
- M. H. Sedri, G. Niedbała, E. Roohi, M. Niazian, P. Szulc, H. A. Rahmani and V. Fieziasl, Agronomy, 2022, 12(7), 1524 CrossRef CAS.
- N. A. Halim, S. B. A. Razak, N. Simbak and C. T. Seng, S. Afr. J. Bot., 2017, 112, 89–94 CrossRef CAS.
- N. M. Zain, M. C. Mat and C. T. Seng, Plant Omics, 2021, 30–37 CAS.
- T. S. Chuah, M. Z. Norhafizah and B. S. Ismail, Crop Pasture Sci., 2015, 66, 214 CrossRef CAS.
- M. Scott, M. Rani, J. Samsatly, J. B. Charron and S. Jabaji, Can. J. Microbiol., 2018, 64, 664–680 CrossRef CAS PubMed.
- P. Cockson, M. Schroeder-Moreno, P. Veazie, G. Barajas, D. Logan, M. Davis and B. E. Whipker, Appl. Sci., 2020, 10, 7875 CrossRef CAS.
- I. A. Atoloye, I. Adesina, A. Shahbazi and A. Bhowmik, Open Agric., 2022, 7, 373–381 CrossRef.
- D. Caplan, M. Dixon and Y. Zheng, Hortscience, 2017, 52, 1307–1312 CAS.
- J. Gorelick and N. Bernstein, in Cannabis sativa L. - Botany and Biotechnology, Springer International Publishing, Cham, 2017, pp. 439–456 Search PubMed.
- M. Brafford May and A. Glode, Cancer Manage. Res., 2016, 49 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3en00380a |
| This journal is © The Royal Society of Chemistry 2023 |