Open Access Article
Open Access ArticleDeciphering silver nanoparticle fate in liver up to biliary excretion using HepG2/C3A spheroids in scenarios mimicking different exposure pathways†
Yousr
Rekik‡
ab,
Vanessa
Tardillo Suárez‡
c,
Vikas Raj
Sharma
ab,
Mireille
Chevallet
a,
Benoit
Gallet
d,
Denis
Falconet
 e,
Peggy
Charbonnier
a,
Isabelle
Kieffer
e,
Peggy
Charbonnier
a,
Isabelle
Kieffer
 f,
Rémi
Tucoulou
c,
Pierre-Henri
Jouneau
*b,
Giulia
Veronesi
f,
Rémi
Tucoulou
c,
Pierre-Henri
Jouneau
*b,
Giulia
Veronesi
 *ac and
Aurélien
Deniaud
*ac and
Aurélien
Deniaud
 *a
*a
aUniv. Grenoble Alpes, CNRS, CEA, IRIG, Laboratoire de Chimie et Biologie des Métaux, F-38000 Grenoble, France. E-mail: giulia.veronesi@cea.fr; aurelien.deniaud@cea.fr
bUniv. Grenoble Alpes, CEA, IRIG, MEM, F-38000 Grenoble, France. E-mail: pierre-henri.jouneau@cea.fr
cESRF, The European Synchrotron, 71 Avenue des Martyrs, 38000 Grenoble, France
dUniv. Grenoble Alpes CEA, CNRS, Institut de Biologie Structurale, 71 Avenue des Martyrs, F-38042 Grenoble, France
eUniv. Grenoble Alpes, CNRS, CEA, INRAE, IRIG-LPCV, Grenoble, France. E-mail: denis.falconet@gmail.com
fBM30 FAME beamline, ESRF, Observatoire des Sciences de l'Univers de Grenoble, Grenoble, France
First published on 23rd May 2023
Abstract
Silver nanoparticles (AgNP) are heavily used in consumer products and medical devices, including face masks to fight against COVID-19. AgNP effects on mammals have been studied both using cellular and animal models. The former elucidate molecular mechanisms occurring at the subcellular level, while the latter bring insight into the toxicity and accumulation at the organ level. It has not yet been possible to reconcile these two kinds of studies in a translational approach. In the current study, we use 3D HepG2/C3A hepatocyte cultures that partially reproduce liver architecture, leading to the formation of active bile canaliculi and exposure scenarios mimicking parenteral or oral exposure to AgNP. By using a combination of cutting edge imaging techniques, atomic spectroscopy, and cell biology, we reveal the formation of Ag sulphide or Ag–organothiol complexes in the different conditions, the storage of excess Ag into vacuoles, the excretion of Ag(I) ions into bile canaliculi through the ATP7B Cu transporter, and modifications of the mitochondrial network. These findings, made possible by the use of a pseudo-organ as a model, reveal exposure-dependent mechanisms that can inspire future studies on nanomaterial metabolism in mammals and risk-assessment, and thereof regulations. Moreover, they can help bridge the gap between cellular and animal studies, bringing significant advances into the understanding of AgNP fate in mammals.
Environmental significanceSilver nanoparticles (AgNP) are massively used in consumer products and medical devices for their biocidal activity, which is due to the release of Ag(I) species. Moreover, AgNP can be transformed into various Ag species in biological and environmental conditions leading to increased exposure to these species and thereof to toxicological and ecotoxicological issues. In human, the main exposure routes are oral and parenteral and silver accumulates in the liver. In this study, the use of innovative 3D hepatic model in conditions mimicking oral and parenteral exposure to AgNP enabled to decipher the fate of Ag species up to their excretion. Three-dimensional cellular models thus offer the opportunity to mind the gap between cell cultures and animal models. |
Introduction
The use of nanotechnologies has been exponentially growing since the nineties, as their incorporation within many materials can offer innovative properties. Among the most frequently found nanomaterials, silver nanoparticles (AgNP) are used for their biocidal activity, which is due to the long-lasting release of the Ag(I) ions produced by oxidation of surface Ag(0) atoms in aerobic conditions.1 Due to their biocidal activity, AgNP are incorporated in various consumer products, including face masks to protect against COVID-19, and in medical devices, to prevent nosocomial infections. Therefore, humans are exposed to Ag species through two major routes: oral and parenteral. It has been estimated that we ingest between 1 and 80 μg of Ag every day,2 whereas little data are available on the human exposure through the parenteral route. Nonetheless, a micromolar concentration of Ag has been measured in the blood of burned patients treated with the Acticoat™ dressings,3 and a recent study described the accumulation of Ag in the liver of patients under long-term hospitalization.4 Data from in vitro studies showed that AgNP are heavily transformed in the gastro-intestinal tract, leading to the translocation of soluble Ag(I) complexes as silver thiolate, or of the AgCl salt, and not of pristine AgNP particles to the blood.5,6 Therefore, the bioavailable species are Ag(I) and AgNP upon oral and parenteral exposure, respectively. This leads to exposure route-dependent metabolism kinetics, as observed in rodents, upon oral exposure, the level in the blood is ten times lower, and in the liver it is 100 to 1000 times lower than upon parenteral exposure.7 Similarly, the clearance rate is much faster in the former exposure scenario than in the latter, 24 h vs. more than 4 days.7 These data are consistent with the presence of different Ag species in the bloodstream: Ag(0) of pristine AgNP and Ag(I) complexes or particles originating from their transformation, in the parenteral and oral exposure route, respectively. This putatively results in different metabolism mechanisms, hence of toxicity thresholds, in the different exposure conditions. For Ag(I) ions, it has already been shown that they can be transported by copper transporters such as Ctr1 and ATP7B,8,9 and excess Cu(I) is known to be excreted via biliary pathway.10 Therefore, unravelling the fate of AgNP in the liver and the excretion mechanisms in these different exposure conditions is of capital importance to evaluate the toxicity of AgNP and define a regulatory framework for their safe use. Nonetheless, the details of AgNP transformations in an animal model are elusive, which originates a gap with conventional cellular studies that offer the possibility to follow particles and their by-products using different types of microscopies. This work aims to fill this gap by applying subcellular imaging and atomic spectroscopy to a physiologically relevant cellular model, in order to disclose physico-chemical transformations that are likely to occur in vivo.In biological media, the surface of nanoparticles gets decorated with proteins and biomolecules that form the so called corona, which defines the biological identity of the nanoparticles, influencing their cellular uptake.11 The relationship between the protein corona forming around PVP- and citrate-coated AgNP and their interactions with different cell types has been recently reviewed.12 Following endocytosis, AgNP are transformed within endolysosomes and release Ag(I) ions that form complexes mainly with organothiol molecules.13–17 State of the art X-ray fluorescence nano-imaging (XRF) coupled with electron microscopy (EM), allowed us to visualize the translocation of Ag(I) complexes to the nucleus and mitochondria of hepatocytes exposed to AgNP.18,19
As of today, it is clear that advanced imaging methods enable to nail down the fate of AgNP in 2D cell cultures. However, considering that 2D cell cultures do not reproduce the physiology of the corresponding organ, it is highly desirable to implement these methodologies in biological models closer to in vivo. For this purpose, we focused this study on 3D hepatic cultures exposed to different Ag species. The kinetics of AgNP and Ag(I) ion uptake and excretion in this model reproduces the trends observed in vivo, proving that the 3D hepatic culture mimics the liver. The combined use of elemental imaging, 3D electron microscopy and speciation analysis showed the partial transformation of AgNP and/or organothiol-Ag(I) complexes into inorganic Ag2S particles, most probably within vacuoles. The excretion of Ag(I) complexes into bile canaliculi was also observed. Moreover, 3D electron microscopy enabled to visualize alterations of the mitochondrial network upon exposure to different types of Ag species. Altogether, by making use of an advanced in vitro model, this work provides insights into the fate of Ag species and the consequent biological effects in the liver of mammals exposed to AgNP via the oral or parenteral pathway.
Methods
Spheroid preparation and exposure conditions
The cells used for the preparation of spheroids, HepG2/C3A, were provided by the ATCC. Spheroids were obtained in two steps. First, freshly trypsinized HepG2/C3A cells were seeded in cell carrier 96-well ULA plate from Perkin-Elmer with 200 cells in 150 μL of media (MEM supplemented with 10% fetal bovine serum (FBS)) per well. After 96 hours of incubation, exposure to Ag species was initiated. Spheroids were exposed to the nominal Ag concentration of 25 μM for cit-AgNP (20 nm Citrate BioPure™ Silver from Nanocomposix), 50 μM for PVP-AgNP (90 nm from Sigma-Aldrich) or 5 μM for AgNO3 (Sigma-Aldrich) for 2, 4 or 7 days with media renewal every 2 days. All silver species were directly added to complete cell culture media. In addition, an excretion protocol was used corresponding to an exposure to the different Ag species for 4 days followed by 3 days without Ag species in the media and renewal every day. Both types of AgNP were previously characterized.15,18 At the end of the exposure procedure, spheroids were rinsed three times in PBS and used for further experiments.For the analysis of bile canaliculi excretion pathway, the excretion protocol was also used. To assess MDR and Mrp pathways, HepG2/C3A spheroids were exposed for 4 days to 50 μM PVP-AgNP and during the 7 days to 50 μM verapamil or 50 μM MK571, respectively. To assess the possible involvement of ATP7B, spheroids made of ATP7B-KO HepG2/C3A cells were prepared and used as for normal HepG2/C3A cells.
Ag quantification by ICP-AES
The amount of Ag present in spheroids was quantified by ICP-AES using a Shimadzu ICPE-9800 after an overnight mineralization of the spheroids at 50 °C in 65% HNO3. Ag amount was normalized based on the number of spheroids per analyzed sample.Sample preparation for XRF, FIB-SEM and STEM-EDX
For high pressure freezing, 5 to 10 spheroids were dispersed in 2 μL of culture media supplemented with 20% FBS and loaded on the 200 μm side of a type A 3 mm gold platelet (Leica Microsystems), covered with the flat side of a type B 3 mm aluminum platelet (Leica Microsystems), and were vitrified by high-pressure freezing using an HPM100 system (Leica Microsystems). Next, the samples were freeze substituted at −90 °C for 40 h in acetone supplemented with 1% OsO4 and warmed up slowly (1 °C h−1) to −60 °C in an automated freeze substitution device (AFS2; Leica Microsystems). After 12 h, the temperature was raised (1 °C h−1) to −30 °C, and the samples were kept at this temperature for another 12 h before a step for 1 h at 0 °C, cooled down to −30 °C and then rinsed four times in pure acetone. The samples were then infiltrated with gradually increasing concentrations of araldite resin in acetone (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 [vol/vol] and pure) for 2 to 8 h while raising the temperature to −10, 0, 10 and 20 °C, respectively. Pure araldite resin was added at room temperature. After polymerization 48 h at 60 °C, 200 nm sections were obtained using an ultra-microtome UC7 (Leica Microsystems) and an Ultra 35° diamond knife (DiATOME) and collected on 50 nm-thick Si3N4 membranes (Silson Ltd) before use at the synchrotron. The same preparation protocol was used for STEM-EDX but the sections were collected on formvar carbon coated 100 mesh copper grids and the sample block was then used in FIB-SEM experiments.
1 [vol/vol] and pure) for 2 to 8 h while raising the temperature to −10, 0, 10 and 20 °C, respectively. Pure araldite resin was added at room temperature. After polymerization 48 h at 60 °C, 200 nm sections were obtained using an ultra-microtome UC7 (Leica Microsystems) and an Ultra 35° diamond knife (DiATOME) and collected on 50 nm-thick Si3N4 membranes (Silson Ltd) before use at the synchrotron. The same preparation protocol was used for STEM-EDX but the sections were collected on formvar carbon coated 100 mesh copper grids and the sample block was then used in FIB-SEM experiments.
Nano-XRF data acquisition and analysis
The data were collected on the hard X-ray nanoprobe beamline ID16B-NA of the European Synchrotron Radiation Facility (ESRF, Grenoble, France). The experiment was performed in air at room temperature. The incoming photon beam energy was set to 29.6 keV. The beam was focused to 65 × 60 nm2 (V × H) with Kirkpatrick–Baez mirrors, providing a photon flux of ∼1011 photons per s. The X-ray fluorescence emission from the sample was recorded using two 3-element silicon drift detector arrays positioned at 13° from the sample. 200 nm-thick sections of spheroids laid on silicon nitride membranes were rapidly scanned at low resolution (1 × 1 μm2 step size, 50 ms per point dwell time) in order to locate the regions of interests. Then, high-resolution images were acquired by scanning the selected areas with 100 × 100 nm2 step size and 500 ms per point dwell time. The hyperspectral images were analyzed using the PyMCA software package (https://pymca.sourceforge.net/).20 The detector response was calibrated over a thin film multilayer sample from AXO (RF8-200-S2453). XRF data were energy calibrated, normalized by the incoming photon flux, and batch-fitted in order to extract spatially resolved elemental concentrations, assuming a biological matrix of light elements and density 1 g cm−3 according to NIST standards (https://physics.nist.gov/cgi-bin/Star/compos.pl?matno=261).XAS data acquisition and analysis
The X-ray absorption spectroscopy experiment was performed in cryogenic conditions on the beamline CRG-FAME-BM30 of the ESRF.21 100 μL drops of spheroid cultures were deposited into a sample holder sealed with Kapton tape and immediately frozen in liquid nitrogen, then transferred into the He cryostat of the beamline and measured at 15 K. The edge region was scanned between 25.30 and 26.48 keV with a nitrogen-cooled Si(220) double-crystal monochromator.22 The incoming photon energy was calibrated with a Ag metallic foil, by defining the first inflexion point of its X-ray absorption spectrum at 25.514 keV. The preparation and measurement of the reference compounds is described elsewhere.23,24 The spectra are deposited in the SSHADE database25 (AgS2OT doi: https://doi.org/10.26302/SSHADE/EXPERIMENT_GV_20181121_001; AgS3OT doi: https://doi.org/10.26302/SSHADE/EXPERIMENT_GV_20181121_002; Ag2S doi: https://doi.org/10.26302/SSHADE/EXPERIMENT_GS_20170712_003). The spectra of cells were recorded in fluorescence mode, with a 30-element Ge solid state detector (Canberra). The data were normalized with standard methods and analyzed as linear combinations of reference compounds by using the Athena software provided with Demeter 0.9.26.26FIB-SEM acquisition
FIB-SEM experiments were performed using a Zeiss crossbeam 550 microscope. In a first step, the resin-embedded sample was coated by a thin layer of Pt (few nm) to avoid charge accumulation. The surface of the sample to be analyzed was then protected by the deposition of 2 μm-thick Pt or C layer on a surface of typically 20 to 50 × 30 μm2. A large hole was then excavated with a Ga+ beam of very high current of 30 nA approximately. The surface of this hole was then trenched slice by slice with the Ga+ beam (with a current of 0.7 or 1.5 nA at a beam acceleration of 30 kV). Each new surface was then imaged with the electron beam. This was done at a low accelerating voltage (around 1.5 kV) and at a current of about 1 nA (except for the imaging of the PVP-AgNP sample which was at I = 4 nA) and using the in-lens backscattered detector (EsB detector, with a bias of 400 V or 600 V). Acquisitions were done at a resolution of 10 nm to obtain a voxel size of 10 × 10 × 10 nm3 except for the AgNO3 sample for which the voxel size was 12 × 12 × 6 nm3. For this sample, the image stack was rescaled afterwards to form 12 × 12 × 12 nm3 isotropic voxels.FIB-SEM analysis
Images were acquired as a series of 2-dimensional TIFF files. In order to compile a 3D tiff file format, the images were first registered in Fiji27 using MultiStackReg28 or Scale Invariant Feature Transform (SIFT) plugins.29 MultiStackReg aligns each image with respect to the previous one using cross-correlation and SIFT is based on the detection and matching of local image features between successive slices. For MultiStackReg, the first or the central image was used as a global reference for the whole stack and translation was used as the transformation type. After alignment, the image stack was checked in orthogonal views and if a misalignment persists, the angle of misalignment was corrected with a python script. The stack of images was then inverted (initial images were acquired in dark field regime), binned and denoised. Anisotropic diffusion30 or median filters served as nice tools to remove noise while preserving organelle membranes. The median filter selects the median gray value of a voxel neighborhood and the anisotropic diffusion filter enables image smoothing with preservation of cell membranes. For some stacks, additional preprocessing steps were performed such as filtering with a variational stationary noise removal filter in Fiji,31 correcting the z drift using the FIB milling position or homogenizing the contrast in the z direction with python scripts.Segmentation of different objects (nuclei, vesicles, vacuoles, Ag-containing particles, mitochondria and bile canaliculi) was done with Ilastik software.32 When needed, manual correction of Ilastik segmentation masks was performed in Microscopy Image Browser.33 The two nuclei of the AgNO3 sample were segmented manually with Imaris (Oxford Instruments). Meshes were either exported from Ilastik or generated in Amira-Avizo (Thermo Fisher Scientific). Visualization and 3D rendering were carried out in Paraview.34
Mitochondria sphericity calculation
Mitochondria segmentation masks exported from Ilastik were imported in Fiji for sphericity calculations. 3D object counter (3D-OC) plugin35 was first used to extract an object map of all mitochondria in each stack based on a voxel connectivity process. Then, the sphericity of all mitochondria found by 3D-OC was calculated by Morpholibj plugin.36STEM-EDX data acquisition and analysis
For the chemical identification of Ag-containing particles, thin sections were observed in STEM mode and analyzed by EDX using either a FEI/Tecnai Osiris microscope or a FEI Titan microscope operated at 200 kV.Results and discussion
Biological model and exposure scenarios
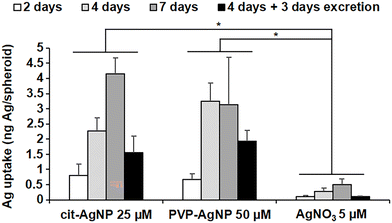
The great majority of nanotoxicology studies made use of either 2D cell cultures or animal models. The former enable the assessment of the disrupted cellular pathways and the intracellular physicochemical transformations of the nanomaterials, while the latter provide an overview of the organismal fate and excretion of the nanomaterials, and their general toxicity. Some studies have used 3D cultures, either to assess the general toxicity,37 or to follow the fate of nanomaterials upon long-term exposure.38,39 However, none of the 3D cell systems used in these studies reproduced the liver metabolism, from the entry up to the excretion of nanomaterials. Recently, we described a 3D hepatocyte model made of HepG2/C3A cells (Fig. S1†).40 In this model, polarized hepatocytes actively excrete organics and inorganics in functional bile canaliculi-like regions, thus mimicking efficiently the liver metabolism. Three dimensional electron microscopy (3D-EM) deciphered the structure of these regions, made of the plasma membranes of neighboring hepatocytes with a high-density of microvilli extensions. Therefore, we used this liver model to analyze the subcellular fate of AgNP and of a Ag salt, up to the excretion of Ag species. Knowing that the transformations of the AgNP depend on their exposure route, we chose different exposure scenarios: to pristine AgNP and to Ag(I) ions, representing parenteral and oral exposure, respectively. As pristine NP, we selected two commonly used AgNP with different coatings that confer them different behaviors in solution, citrate-coated AgNP of 20 nm in diameter (cit-AgNP) and PVP-coated AgNP of 90 nm in diameter (PVP-AgNP). In cell culture media, PVP-AgNP are covered by a protein corona and remain monodisperse with a diameter around 90 nm, while cit-AgNP form aggregates of about 160 nm, as we previously observed.15,18 In addition, AgNO3 was used to expose the cells directly to dissolved Ag(I) ions. Besides, all exposures were performed in cell culture media supplemented with FBS that is the closest condition to mimic the bloodstream. AgNP can thus be considered as covered by a protein corona under the chosen exposure conditions. 3D hepatocyte cultures were exposed to the Ag species for 2, 4 or 7 days, or for 4 days followed by 3 days without exposure to favor their excretion (excretion scenario). The goal was to analyze over several days the uptake, the transformation and the excretion of AgNP and Ag(I) ions in the liver that is the main target in humans, regardless of the exposure route. Previous experiments using 2D cultures showed that the first signs of toxicity for cit-AgNP, PVP-AgNP and AgNO3 appear at 50, 100 and 5 μM, respectively.15 However, cytotoxicity is observed at higher doses in 3D compared to 2D cultures. Therefore, in order to fully preserve the viability of the cells while exposing them to a significant amount of Ag, we chose to work at 25, 50 and 5 μM in Ag for cit-AgNP, PVP-AgNP and AgNO3, respectively. To analyze Ag uptake and its balance with excretion, the amount of Ag per spheroid was quantified by inductively coupled plasma atomic emission spectroscopy (ICP-AES) (Fig. 1). For cit-AgNP and AgNO3, the amount of Ag gradually increased from 2 to 7 days of exposure, while for PVP-AgNP it reached a plateau after 4 days. However, for both types of AgNP, the maximum level of internalized Ag was similar, between 3 and 4 ng Ag per spheroid. The Ag accumulation with AgNO3 was 6 to 8 times lower than with AgNP. Moreover, the excretion scenario showed a slower decrease in Ag in the case of AgNP exposure compared to a Ag salt. Indeed, after 3 days of excretion, the fraction of excreted Ag was 65% in the case of AgNO3, and between 30 and 40% for the two types of AgNP. This means that the dynamic for uptake, transformation, trafficking and excretion is faster upon exposure to dissolved Ag(I) compared to AgNP, which are a mixture of Ag(0) crystals covered by a Ag(I) layer. These data are consistent with experiments performed in mice exposed orally or intravenously to AgNP which showed a fast decrease of Ag in blood and liver in the former case, while Ag remains high for several days in the latter case.7 Taken together, these results confirm the relevance of the 3D hepatocyte model to mimic the liver, and the consistency of the chosen exposure scenarios to mimic oral and parenteral exposure to AgNP.In the last years, we have shown the interest of multimodal imaging, including XRF and EM, to analyze, with the highest sensitivity and resolution, the fate of AgNP in 2D cell cultures.15,18,19 In this study, we improved the methodology and extend it to a 3D hepatocyte model. First, the samples were prepared by high pressure freezing followed by cryo-substitution using acetone and not ethanol, in order to preserve the cell ultrastructure as well as its elemental content. Second, 200 nm-thick cell sections were analyzed by XRF at 100 nm2 resolution, and 3D reconstructions from the remaining sample block were obtained at nanometer resolution with focused ion beam scanning electron microscopy (FIB-SEM). This was complemented by scanning transmission electron microscopy coupled with energy dispersive X-ray spectroscopy (STEM-EDX). Finally, bulk speciation analyses from 3D hepatocyte cultures were performed by X-ray absorption spectroscopy (XAS) to obtain quantitative information on Ag speciation under the selected conditions.
Trafficking and transformations of AgNP in 3D hepatocyte cultures
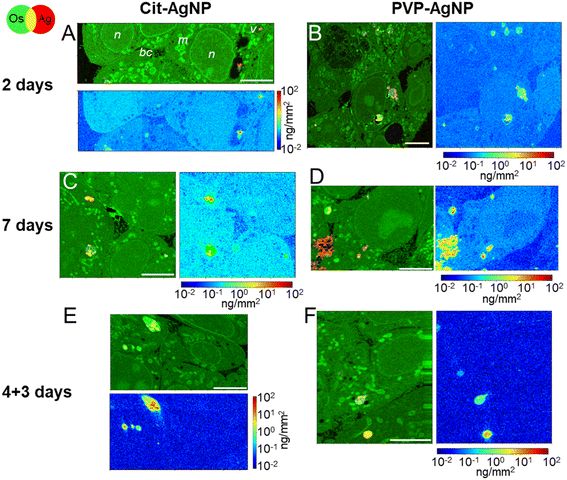
The internalization of AgNP in hepatocytes, their progressive dissolution and trafficking up to their excretion was followed by means of XRF imaging and EM. With this multimodal imaging approach, we meant to retrieve the distribution of silver, regardless of its chemical state, in the different subcellular compartments, and, when excretion takes place, in bile canaliculi. Selected elemental images extracted from the XRF hyperspectral images collected in a synchrotron nanoprobe are reported in Fig. 2. All other images collected on 3D hepatocyte cultures exposed to cit-AgNP and PVP-AgNP are reported in Fig. S2 and S3,† respectively. For each exposure condition, we acquired at least three high-resolution images containing several cells, and, whenever possible, bile canaliculi. For all datasets, two maps representing the same sample area are reported (see Fig. 2A as an example): a first map shows the localization of Os (osmium, green) and Ag (red). The presence of Os is due to the use of OsO4 for sample preparation, which reacts with lipids. Os signal thus reveals the structural details of the interior of the cell with high resolution, allowing for the identification of nuclei (n), mitochondrial networks (m), vesicles (v), and bile canaliculi (bc). The latter are recognizable as intercellular spaces filled by villi. Vesicles are often loaded with Ag (red signal), clearly indicative of the presence of nanoparticles internalized through endocytosis. Next to each Os and Ag co-localization map, the concentration of Ag in the same map area is reported. The concentrations are represented as areal densities ranging over five orders of magnitude, from 0.01 to 100 ng mm−2, on a logarithmic scale. This visualization reveals the presence of Ag not only as highly concentrated hot-spots (10–100 ng mm−2) corresponding to NPs in vesicles, but also as a much weaker signal of the order of 0.05–0.1 ng mm−2. A value of 0.05 ng mm−2 corresponds to 0.6 × 10−18 g in the area probed by the beam, which gives an empirical estimation of the detection limit for Ag in our experimental conditions. In our previous work, we made a synergistic use of XAS, STEM/EDX, and XRF, which allowed us to assign the weak Ag signal observable all over the cell area in XRF to Ag(I) ions that are released from the NPs in vesicles and diffuse in the cytosol, where they form molecular complexes with thiolate biomolecules.18,19 In Fig. 2, the concentration of Ag can be visually compared among the different conditions, since the scale is the same for all maps.After 2 days of exposure to cit-AgNP (Fig. 2A) or to PVP-AgNP (Fig. 2B), the NPs have partially dissolved in the endolysosomes, as clearly visible in Fig. 2B, for instance, and Ag(I) complexes are found in all cell compartments such as cytoplasm and nuclei. The concentration of Ag in the cell area appears to increase with the exposure time, for both kind of NP (Fig. 2Avs. C and B vs. D). This can be observed as a general trend among all Ag concentration maps, despite the variability between one cell and another (Fig. S2 and S3†). Interestingly, after 7 days of exposure to PVP-AgNP, Ag is clearly observed in some bile canaliculi (Fig. 2D), with a spatial distribution that differs markedly from the hot-spots observed in the presence of nanoparticles, with its concentration roughly constant over bile canaliculi area and about one order of magnitude lower than NP found in endosomes. After four days of exposure to cit-AgNP, we observed the presence of large (∼3–4 μm diameter) empty vesicles, which could putatively be vacuoles that store excess Ag, probably to protect the cell (Fig. S2C†). Interestingly, this feature has never been observed in 2D cultures, neither by us in hepatocytes, nor, to the best of our knowledge, in other cell types.
Finally, in the excretion scenario, i.e. AgNP exposure is stopped after four days, and then the cells are left to grow for three more days and rinsed daily with fresh media, Ag is observed in vesicles but not or only barely in the remaining cell area (Fig. 2E and F and S2F and G and S3B and C†). This implies that the Ag(I) complexes, observed in the cells already after two days of exposure, were largely excreted. This observation corroborates the hypothesis of the preferential excretion of Ag(I) soluble species with respect to the crystalline Ag(0) form or to transformed AgNP. In order to obtain more independent evidence for this phenomenon, we performed electron microscopy experiments. In the first instance, spheroid sections were observed by STEM-EDX, for samples exposed to AgNP, which showed neither electron-dense material, nor a Ag signal by EDX in bile canaliculi areas (Fig. S4†). However, it is possible to miss electron-dense objects by using cell sections that lead to a limited volume of analysis. To improve this point, FIB-SEM acquisitions were performed on samples that had been exposed for 7 days to both types of AgNP (Fig. 3, and Videos S1 and S2†). FIB-SEM allows the acquisition of volumes of several thousands of μm3. In these stacks of EM images, electron dense objects were never observed in bile canaliculi regions. For cit-AgNP, vacuoles with large amounts of AgNP and transformed AgNP were observed (Fig. 3B and C and S5†). An event of a fusion between an endolysosome and a vacuole was also observed (Fig. 3B and C). Therefore, in the case of cit-AgNP exposure, it is clear that to cope with the huge accumulation of cit-AgNP, the fast dissolution into Ag(I) ions and the slow excretion of the Ag species in the hepatocytes in 3D culture system, cells exposed to cit-AgNP store Ag particles inside vacuoles. This phenomenon was not observed in more classical 2D hepatocyte cultures, in which the excretion occurs in the large interface between cell plasma membranes and cell culture media. 3D cultures are thus better suited to mimic the fate of NP in the liver. Moreover, the fate and excretion could be different upon exposure to PVP- or cit-AgNP. Indeed, XRF showed Ag species in bile canaliculi areas only for the former, while vacuole storage was only observed for the latter. Currently, we are not able to provide an explanation for this difference, but we hypothesize that the intracellular concentration of soluble Ag(I) complexes reaches a threshold after which hepatocytes have to store Ag-particles in vacuoles to avoid toxicity, and that this threshold is reached only by cit-AgNP. In support of this hypothesis, we observed that upon cit-AgNP exposure, the excretion of Ag is the slowest showing only 30% of Ag excreted outside the spheroids after 3 days (Fig. 1).
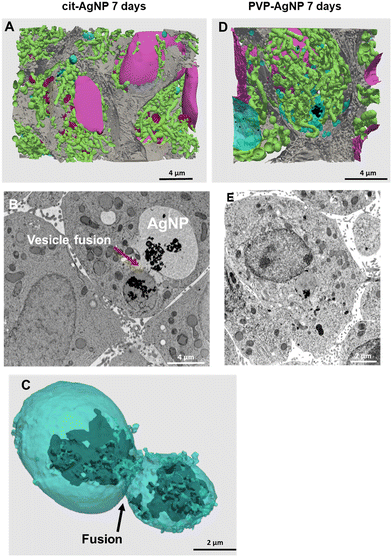 | ||
| Fig. 3 3D reconstruction and segmentation of spheroids exposed to AgNP. (A) From the stack of images shown in Video S1,† the inside of a spheroid exposed to cit-AgNP for 7 days was reconstructed, segmented in the different organelles and represented with nuclei (pink), mitochondria (green), vesicles (blue), bile canaliculi (grey) and Ag-containing particles (black). The segmentation and 3D rendering are shown in Video S1.† (B) Electron micrograph of a single XY slice showing the fusion between a vacuole and a vesicle. (C) 3D rendering of the fusion of these vesicle and vacuoles. (D) From the stack of images shown in Video S2,† the inside of a spheroid exposed to PVP-AgNP for 7 days was reconstructed, segmented in the different organelles and represented with nuclei (pink), mitochondria (green), vesicles (blue), bile canaliculi (grey) and Ag-containing particles (black). The segmentation and 3D rendering are shown in Video S2.† (E) Electron micrograph of a single XY slice showing a AgNP-containing vesicle bound to the nuclear membrane. | ||
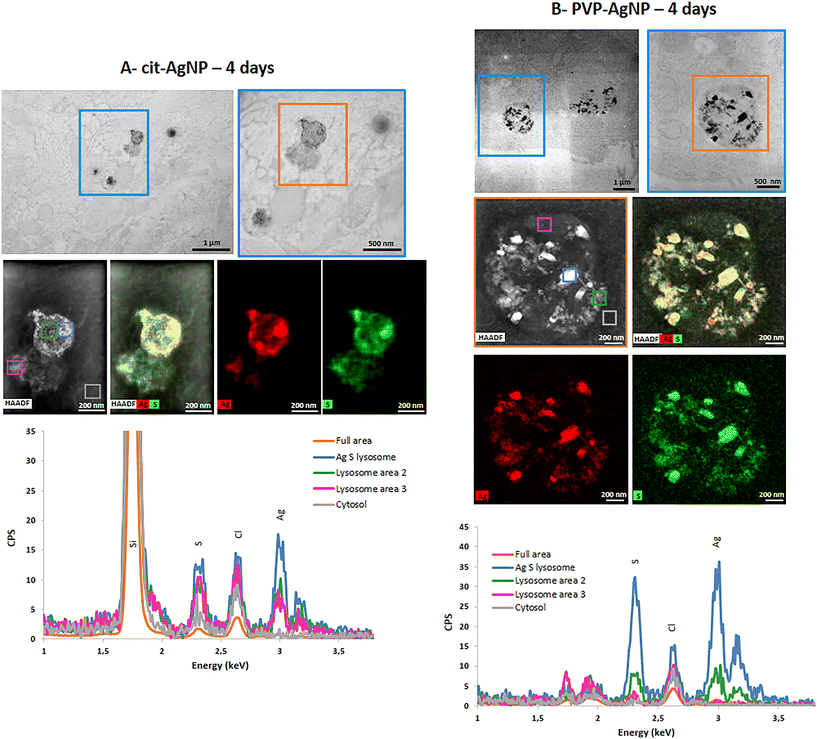
STEM-EDX analysis of the elemental content of different Ag-particles, upon transformation in vesicles or vacuoles, always showed the co-localization of Ag and S with similar peak intensity, while chloride peak had a similar intensity to the level observed in non-particle locations (Fig. 4A and B and S5 and S6†). Therefore, whatever the type of vesicles and coatings, AgNP are transformed into AgS complexes/particles with different shapes including sheets (Fig. S6†), small round particles (Fig. 4A) or larger, irregular objects (Fig. 4B).
Trafficking and transformations of AgNO3 in 3D hepatocyte cultures
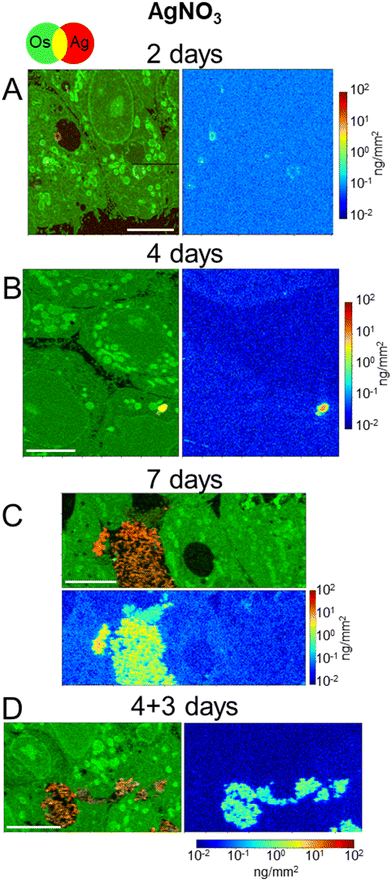
In order to discern the different cellular mechanisms that rule the detoxification from nanoparticles or from ions, we performed XRF and EM experiments on 3D hepatocyte cultures that were exposed to the AgNO3 salt under the same exposure scenarios applied to AgNP. This allowed us to observe that the trafficking and excretion processes operated by hepatocytes were faster when the cells were exposed to the Ag salt rather than to AgNP. A sub-lethal dose of 5 μM AgNO3 was administered, for 2, 4, 7 days, or in the excretion scenario (4 + 3 days). One XRF image per exposure condition is reported in Fig. 5. All other images, collected to increase the statistical significance of the results, are reported in Fig. S7.† The visualization is the same as for NP-exposed samples, including the range of the Ag concentration scale, to ease the comparison between different exposure conditions. Os (green) and Ag (red) co-localization maps in Fig. 5 and S7† reveal the presence of the latter in vesicles (e.g. in Fig. 5B), vacuoles (e.g. in Fig. 5A and S7E and F†), and very often in bile canaliculi (Fig. 5C and D and S7C and G†). A faint Ag signal is observed in the cell area after four or seven days of exposure (Fig. 5B and C and S7A–D†), where it is slightly distinguishable from the background. This signal is never observed in the excretion scenario, where Ag is instead systematically found in the bile canaliculi, suggesting that the excretion via the bile canaliculi targets the free soluble Ag(I) complexes that are not segregated into vesicles or vacuoles. Interestingly, the presence of Ag in bile canaliculi is observed only in the case of a 7 day exposure or in the excretion scenario. For shorter exposure times and exposures with no recovery (2 and 4 days), Ag is observed mainly in vesicles and vacuoles. This is probably due to the increase in Ag content in bile canaliculi over time as well as a higher number of bile canaliculi loaded with Ag that makes it easier to visualize Ag-loaded bile canaliculi 7 days after the beginning of AgNO3 exposure.It can be noticed that, in spite of the lower Ag dose administered in the form of salt with respect to NPs, and of the consequently lower intracellular accumulation of Ag, Ag-loaded bile canaliculi are more frequently observed in cells exposed to AgNO3 than in those exposed to AgNP. A possible explanation would be that the main excretion pathway targets Ag(I) soluble species, which are readily available upon AgNO3 exposure. In contrast, when AgNP are endocytozed into the cells, Ag(I) ions are progressively released from their surface within endolysosomes, limiting the rate of formation of molecular Ag complexes. Electron microscopy was performed to complete the understanding of the fate of AgNO3 in hepatocyte 3D cultures. As for AgNP exposure, FIB-SEM and STEM-EDX did not show electron-dense particles nor was a Ag signal detected by EDX in bile canaliculi areas, respectively (Fig. 6A and B and S8, and Video S3†). These data suggest that Ag(I) complexes are excreted in bile canaliculi. However, electron-dense materials were observed by EM in vesicles and vacuoles (Fig. 6C and D ) and Ag was detected by EDX (Fig. S9 and S10†) proving that Ag hot spots observed by XRF corresponds to Ag particles. Moreover, EDX analyses of these particles showed the co-localization of S and Ag in the vesicles and vacuoles with a similar intensity for both peaks, as observed with AgNP-exposed sample analyses. These data highlight the point that after exposure to a Ag salt, the Ag(I) ion recombines with S to form AgS particles within these specific organelles in the cell. This is consistent with previous data obtained in rodents exposed to a Ag salt and in which Ag particles were detected in the liver.41,42 A 3D hepatocyte culture model is thus particularly relevant to represent the fate of different kinds of Ag species in mammal liver. However, the speciation of Ag in these particles remains unknown up to now, making the comparison of the Ag salt versus AgNP fate in this biological system of high interest. To go further, we performed XAS analysis on the bulk spheroids to understand the transformations of Ag species in hepatocytes grown in 3D.
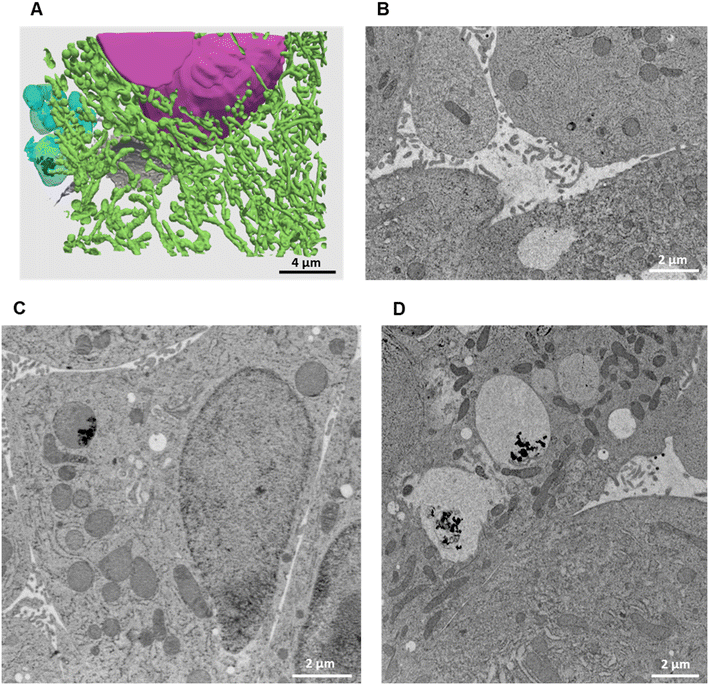 | ||
| Fig. 6 3D reconstruction and segmentation of spheroids exposed to AgNO3. (A) From the stack of images shown in Video S3,† the inside of a spheroid exposed to AgNO3 for 7 days was reconstructed, segmented in the different organelles and represented with nuclei (pink), mitochondria (green), vesicles (blue), bile canaliculi (grey) and Ag-containing particles (black). The segmentation and 3D rendering are shown in Video S3.† (B) Electron micrograph of a single XY slice showing a bile canaliculi area. (C) Electron micrograph of a single XY slice showing an endolysosome containing Ag-particles. (D) Electron micrograph of a single XY slice showing vacuoles containing Ag-particles. | ||
Speciation analysis to understand AgNP and AgNO3 transformations in 3D hepatocyte cultures
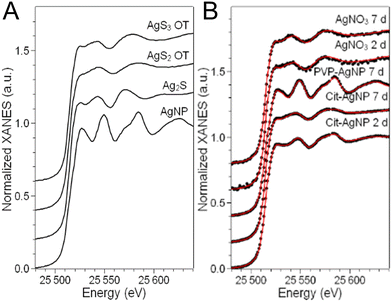
In order to disclose the chemical transformations that AgNP and a Ag salt go through in the hepatocyte 3D model, we performed XAS on the exposed spheroids. The linear combination fitting (LCF) of the near-edge spectral region (X-ray Absorption Near-Edge Structure, XANES) allows the detection of the Ag species present in the specimen and their relative fractions, provided that an exhaustive set of reference compounds is used. As reference compounds that represent the expected Ag species in cellulo, we chose pristine AgNP (i.e. as-synthesized citrate or PVP coated AgNP in colloidal suspension), corresponding to the undissolved fraction of AgNP in cellulo, Ag chloride and Ag–sulfur compounds, which are their commonest transformation products in the environment and in cells. In this study, we made a careful choice of different Ag–sulfur reference compounds in order to finely distinguish the transformation products of AgNP and AgNO3. This allowed us to propose different mechanisms for handling the toxic Ag(I) ions in the two cases. Indeed, Ag–sulfur compounds deserve a special focus: in biological systems exposed to AgNP, the formation of both Ag2S crystals and Ag–organothiol (Ag–OT) compounds has been reported.15,16,43 The latter are, for instance, the complexes formed with GSH and Met, where Ag occupies digonal AgS2 and trigonal AgS3 sites, respectively.24 The spectra of the three Ag–S reference compounds, i.e. the Ag2S crystal and two Ag–OT compounds in AgS2 and AgS3 coordination, reported in Fig. 7A, show small but significant variations depending on the coordination chemistry of Ag. In particular, the second post-edge oscillation, centered at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 570 eV in the spectrum of Ag2S, shifts at higher energy in the two Ag–OT compounds. Therefore, for a reliable LCF analysis, all three Ag–S references must be included. By doing so, and by extending the fitting range up to ∼25
570 eV in the spectrum of Ag2S, shifts at higher energy in the two Ag–OT compounds. Therefore, for a reliable LCF analysis, all three Ag–S references must be included. By doing so, and by extending the fitting range up to ∼25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 eV in such a way that the second post-edge oscillation is included, the LCF of the XANES spectra can provide information about the organic or inorganic nature of the Ag–S complexes formed in spheroids.
600 eV in such a way that the second post-edge oscillation is included, the LCF of the XANES spectra can provide information about the organic or inorganic nature of the Ag–S complexes formed in spheroids.
The LCF analysis results, providing the fraction of each Ag species in cells in the different exposure conditions, are reported in Table 1. The AgCl component was never detected, so the corresponding column is omitted.
| Ag source | Exposure time | XANES LCF results | ||||
|---|---|---|---|---|---|---|
| AgNP (%) | AgS2 OT (%) | AgS3 OT (%) | Ag2S (%) | R fit (10−4) | ||
| Cit-AgNP | 2 d | 26 (1) | 50 (4) | 3 (5) | 21 (4) | 3.3 |
| Cit-AgNP | 7 d | 5 (1) | 75 (4) | — | 20 (2) | 2.2 |
| PVP-AgNP | 7 d | 71 (2) | 22 (5) | — | 7 (3) | 4.1 |
| AgNO3 | 2 d | — | 8 (10) | 78 (7) | 14 (5) | 5.5 |
| AgNO3 | 7 d | — | 22 (4) | 66 (7) | 12 (5) | 5.0 |
We observe that the AgNP fraction in hepatocytes upon two days of exposure to cit-AgNP is 26 ± 1%, confirming that the NPs quickly transform, as observed in 2D cell cultures.15,18 The transformation is complete or almost (5% AgNP, below the detection limit of the technique) after 7 days of exposure to the same NP. For these AgNP, the fraction of Ag(I) occupies both Ag2S crystal sites (∼20% of total Ag, regardless of the exposure time) and digonal Ag–OT sites, notably with GSH but also in various proteins such as in Zn finger-containing proteins as we described recently.18,44,45 PVP-coated NPs show a slower transformation with 71 ± 2% of the Ag atoms still in the Ag(0) state of pristine NPs. The slower transformation of PVP-coated AgNP with respect to citrate-coated AgNP in 2D hepatocyte cultures as well as in other cell cultures has been largely documented. It can be partly due to the larger diameter of this NP, and partly to the different ligands used for stabilization (PVP vs. citrate, used for sterical and electrostatic stabilization, respectively).15,16 No AgNP formation is observed when 3D hepatocyte cell cultures are exposed to Ag(I) ions from the AgNO3 salt (Table 1). Both inorganic and organic Ag(I)–sulfur species form, in proportions that differ significantly from the ones observed in the case of exposure to AgNP. In particular, the amount of inorganic Ag2S is constant within the error (12–14%) between 2 and 7 days of exposure to the salt, and slightly lower than the amount of Ag2S formed upon the complete transformation of cit-AgNP in 7 days (20 ± 2%). Between the Ag–OT species formed, the most relevant geometry is trigonal AgS3, as in Met, whereas digonal AgS2-OT are minor (22 ± 4% after 7 days of exposure) or below the detection limit. This is the main difference observed depending on the origin of the Ag(I) ions released in hepatocytes. The coordination chemistry of the Ag–OT complexes formed in cellulo is different depending if Ag(I) ions are slowly released from AgNP or readily released by a Ag salt. This suggests that the cells set up different mechanisms to handle Ag(I) ions in the two cases. It has been demonstrated that the exposure to AgNP induces Met overexpression in several human cell lines, as Caco-2, NHBE, BEAS-B2 or HepG2 cells,15,46 which is considered as a part of the cellular response to metal-induced stress. This is possible when the Ag(I) are slowly released in cells from the AgNP surface. In the case of direct exposure to a salt, instead, the entry of Ag(I) ions is faster and the Met overexpression mechanism is less efficient, resulting in an enhanced toxicity.
Overall, the main process of AgNP transformation is conserved between 2D and 3D cell cultures which leads to the release of Ag(I) ions that form Ag–OT complexes. However, XAS analyses revealed the significant presence of inorganic Ag2S species in cit-AgNP and AgNO3 conditions, even though we never observed this type of species in 2D hepatocyte culture. One can assume that it corresponds to particle species localized within vacuoles, since we also never observed Ag-containing vacuoles in 2D cell culture. Thereof, vacuoles would provide HS− or H2S species that is a suitable chemical environment for Ag2S formation. This kind of silver sulfidation mechanism has already been described in specific immunocompetent regions of the brain.47 Besides, Ag–OT represent the great majority of the Ag species found in 3D hepatocyte cultures exposed to AgNO3 (Table 1), which suggests that the Ag rich areas visible in the bile canaliculi of exposed cells (Fig. 5 and S7†) contain excreted Ag–OT complexes. These data support our model for Ag(I) ions or molecular complexes excretion in bile canaliculi.
XAS data are also consistent with STEM-EDX mapping, which showed the presence of electron-dense materials in which Ag and S are co-localized in sections of spheroids exposed to cit-AgNP, PVP-AgNP and AgNO3 (Fig. 4 and S5, S6, S9 and S10†). Since the proportion of the different species is very diverse depending on the type of exposure, we have to consider that electron-dense particles observed in endolysosomes are not all the same. In the case of PVP-AgNP and cit-AgNP, these objects are pristine particles and particles being transformed into AgS2 species in different proportions depending on the NP. For AgNO3 exposure, these by-products are Ag–OT and mainly AgS3. However, the process driving the latter to endolysosomes remains elusive.
Molecular pathway for the main excretion of Ag species
Since we found that Ag is excreted as Ag(I) ions or molecular complexes in bile canaliculi, we assessed three possible excretion pathways: (i) via Mrp2 through which GSH-Ag(I) complexes could be transported, (ii) via ATP7B, a Cu(I) transporter, (iii) via MDR pumps that are unlikely to transport Ag(I) species. The excretion exposure scenario was used and applied to PVP-AgNP exposure, since this condition resulted in a significant excretion of Ag species in bile canaliculi, as reported above. In addition, verapamil or MK571 were added to inhibit the excretion via MDR or Mrp2, respectively. To test for the excretion via ATP7B, we used a cell line that was derived from HepG2/C3A from which we deleted its gene by CrispR/Cas9.48Hepatocytes were treated in such a way that the excretion pathways are selectively inhibited. At the chosen time point, cells were prepared and analyzed by nano-XRF. The accumulation of Ag in cells was assessed by measuring its concentration in selected areas of the XRF maps in the cytosol. At least three cells per condition were analyzed and the average Ag areal density in the cytosol of hepatocytes in the different conditions are reported in Table 2. The control samples in this experiment are HepG2/C3A with intact molecular pathways.
| Sample | Treatment | n | Ag in cytosol (ng mm−2) |
|---|---|---|---|
| Control | None | 4 | 0.052 ± 0.012 |
| Mrp2-inhibited | MK571 | 5 | 0.025 ± 0.003 |
| MDR-inhibited | Verapamil | 3 | 0.056 ± 0.010 |
| ATP7B-absent | atp7b gene deletion | 4 | 0.100 ± 0.014 |
We observe that the inhibition of MDR (verapamil) does not influence the accumulation of Ag in hepatocytes. In the case of inhibition of Mrp2 (MK571), the concentration of Ag is significantly lower than in the control cells that present functional Mrp2 pumps. In contrast, deleting the atp7b gene results in a dramatic accumulation of Ag in hepatocytes, double than in unaltered cells. This means that the excretion is drastically inhibited, which in turn proves that Ag(I) is mainly excreted in the bile canaliculi as ions transported through the ATP7B Cu transporter, and not as GSH-Ag(I) complexes through Mrp2. Our data are consistent with a previous publication showing that dissolved Ag from AgNP induced ATP7A trafficking to the plasma membrane of Caco2 cells.49
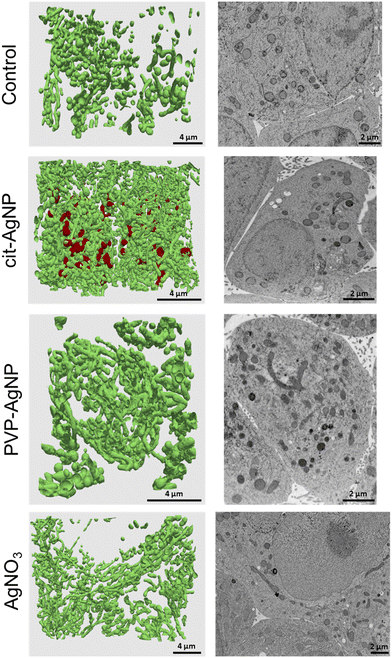
Ultrastructural alterations analyzed by 3D electron microscopy
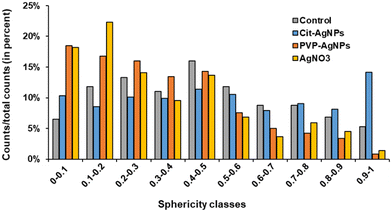
3D hepatocyte cultures exposed for 7 days to cit-AgNP, PVP-AgNP and AgNO3 have been analyzed by FIB-SEM. This approach offers the advantage to acquire a volume of more than 1000 μm3 within the spheroid and at 10 nm resolution (Fig. 3 and 6, and Videos S1–S3†). As shown previously, it allowed us to visualize the distribution of Ag-particles within these volumes and thus provided a more accurate information than thin section analyses. In addition, the raw FIB-SEM volumes were segmented into the different types of organelles or the intercellular region that constitutes the bile canaliculi (Fig. 3 and 6). Among the different organelles, visual inspection showed that the mitochondrial network looks different between the control sample and the hepatocytes exposed to AgNP or AgNO3 (Fig. 8). Indeed, upon exposure to cit-AgNP, it appears that mitochondria at the extremities of the network are swollen (red mitochondria), while upon exposure to PVP-AgNP and AgNO3, mitochondria look more elongated forming tube-like networks. To confirm these observations, quantitative analyses were performed (Fig. 9). The sphericity of all the mitochondria present in each volume were determined. A sphericity value of 1 corresponds to a mitochondrion that would be a perfect sphere. Control sample had a sphericity distribution with mitochondria of various sphericity with a maximum in the distribution for the class between 0.4 and 0.5 in sphericity. Cit-AgNP had an even distribution except for the class 0.9–1 that was about twice more populated than the others. This class can clearly correspond to the swollen mitochondria. PVP-AgNP and AgNO3 samples had similar sphericity distributions that were shifted towards smaller values with the classes between 0 and 0.2 being the most populated. These classes correspond to 34 and 40% of all the mitochondria for PVP-AgNP and AgNO3 exposures, respectively. These results corroborated the visual inspection since it meant that mitochondria were more elongated in these two samples. Therefore, FIB-SEM analyses demonstrated that mitochondria in hepatocyte spheroids are affected upon exposure to AgNP or a Ag salt. We cannot yet clearly establish the reason of the opposite effect observed between cit-AgNP on one side, and PVP-AgNP and AgNO3 on the other side. However, based on XAS and XRF analyses, it is clear that the exposure to cit-AgNP induces the highest intra-cytosolic concentration of Ag(I) molecular species, which could explain the presence of swollen mitochondria as a mark of mitochondrial toxicity. In the case of PVP-AgNP and AgNO3, the amount of soluble Ag(I) in the cytosol is lower and not heavily toxic for mitochondria, but it seems to impact mitochondrial fusion–fission equilibrium, which could be, for instance, due to an adaptation of cellular metabolism in these conditions. These data are consistent with recent publications that described an impact of AgNP exposure on mitochondrial biogenesis and dynamics.50–52 However, this effect would not be directly due to interactions between AgNP and mitochondria but it could be explained by mitochondrial uptake of Ag(I) species that we recently showed to happen upon AgNP exposure.19Conclusions
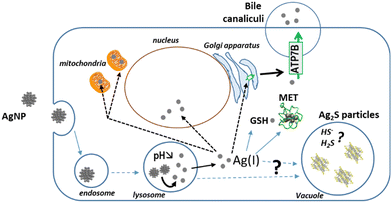
To follow the fate, from entry to excretion, of different types of Ag species in a model mimicking the liver, we developed 3D hepatocyte cell cultures with active bile canaliculi-like structures that perform excretion similar to in vivo. Besides, we combined state-of-the-art synchrotron-based elemental imaging with 3D electron microscopy to visualize all Ag species with a resolution that allowed to locate them at the organelle level. The sample preparation was optimized to preserve the elemental content of the cells. By doing so, we could assess AgNP and AgNO3 transformations in the liver with unequaled details, and unveil the Ag species excretion pathways (Fig. 10). It has been shown that the AgNP exposure pathway influences Ag clearance, which is very slow upon parenteral compared to oral exposure.7 To mimic these two situations, we have exposed spheroids to AgNP or AgNO3, respectively, and observed similar trends in the accumulation and clearance of Ag. According to our results, Ag excretion is mainly mediated by transport of Ag(I) ions to bile canaliculi and through the Cu transporter ATP7B. These results are also consistent with clinical data that revealed an accumulation of Ag in the liver from patients with long-term hospitalization.4 Indeed, in this case the patients were exposed to pristine AgNP via parenteral exposure due to the release of AgNP from catheters. Moreover, in this cohort, Wilson patients with a defective ATP7B protein had significantly higher Ag accumulation compared to the whole cohort. These data are therefore consistent with the main liver excretion pathway we evidenced, i.e. via ATP7B.The imaging methodology also allowed us to analyze the transformations of AgNP into other Ag species within the spheroids (Fig. 10). Ag-containing particles were observed following AgNO3 exposure as shown in the liver of mice exposed to a Ag salt,41,42 confirming the in vivo relevance of our spheroid model. These Ag particles were located both in endolysosomes and vacuoles and correspond to both organothiol complexes (AgS2 and AgS3) and to inorganic Ag2S. Interestingly, this type of inorganic Ag was also detected by XAS upon cit-AgNP exposure. Concomitantly, Ag particles were visualized in vacuoles in this exposure condition, while we never observed Ag particles in vacuoles nor detected Ag2S in 2D hepatocyte culture. Therefore, we formulate the hypothesis that hepatocytes can store Ag excess in vacuoles in the form of Ag2S particles.
Finally, 3D electron microscopy enables to analyze cell ultrastructure in 3D at nanometer resolution. This approach revealed that the exposure to Ag can trigger either mitochondrial swelling, in conditions of high organothiol-Ag(I) accumulation (cit-AgNP), or mitochondrial network remodeling towards a more tubular morphology under low to moderate organothiol-Ag(I) accumulation (PVP-AgNP, AgNO3). The ultrastructure of the other organelles and bile canaliculi were not significantly altered upon Ag exposure.
In conclusion, hepatocyte spheroids appeared to be a useful and pertinent tool to understand the fate of AgNP in the liver, since it recapitulates all data known from animal studies and it revealed information not accessible so far. Thus, this is an outstanding model to shed new lights on the metabolic liver transformations and excretion of engineered nanomaterials, beyond the specific case of AgNP. However, the integration of Kupffer cells in this system would enable to take into account the uptake and transformations performed by these macrophages.
Author contributions
GV and AD conceived the project. PHJ, GV and AD designed the study. YR, VTS, VRS, MC, BG, DF, PC, IK, PHJ, GV and AD performed the experiments. YR, VTS, VRS, IK, RT, PHJ, GV and AD participated in data analysis. GV and AD wrote the manuscript.Conflicts of interest
Authors have no competing interests to declare.Acknowledgements
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities (project LS-2967 on BM30 and LS-2710 on ID16B). This work used the platforms of the Grenoble Instruct-ERIC Center (ISBG: UMS 3518 CNRS-CEA-UGA-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). The EM facility is headed by Guy Schoehn and supported by the Auvergne Rhône-Alpes Region, the Fondation Recherche Medicale (FRM), the fonds FEDER and the GIS-Infrastrutures en Biologie Santé et Agronomie (IBISA). YR PhD is supported by the LabEx Gral and this project also received funding from Arcane Labex, programs from the Chemistry Biology Health (CBH) Graduate School of University Grenoble Alpes (ANR-17-EURE-0003). This work was also supported by the CEA DRF-Impulsion grant FIB-Bio and the Région Auvergne Rhône-Alpes.References
- J. Liu and R. H. Hurt, Ion Release Kinetics and Particle Persistence in Aqueous Nano-Silver Colloids, Environ. Sci. Technol., 2010, 44, 2169–2175 CrossRef CAS PubMed.
- S. W. Wijnhoven, W. J. Peijnenburg, C. A. Herberts, W. I. Hagens, A. G. Oomen, E. H. Heugens, B. Roszek, J. Bisschops, I. Gosens and D. Van De Meent, Nano-silver–a review of available data and knowledge gaps in human and environmental risk assessment, Nanotoxicology, 2009, 3, 109–138 CrossRef CAS.
- E. C. M. Mimura, J. P. M. Favoreto, M. E. Favero, K. L. Bonifacio, T. S. Peixe, A. A. Morita, D. S. Barbosa, M. J. S. Yabe and A. J. F. Carrilho, Silver serum levels in burned patients treated with silver sulfadiazine and its toxicity on inflammatory cells, Burns, 2020, 46(5), 1120–1127 CrossRef PubMed.
- J. Poznański, D. Sołdacki, B. Czarkowska-Pączek, A. Bonna, O. Kornasiewicz, M. Krawczyk, W. Bal and L. Pączek, Cirrhotic Liver of Liver Transplant Recipients Accumulate Silver and Co-Accumulate Copper, Int. J. Mol. Sci., 2021, 22, 1782 CrossRef PubMed.
- P. Bove, M. A. Malvindi, S. S. Kote, R. Bertorelli, M. Summa and S. Sabella, Dissolution test for risk assessment of nanoparticles: a pilot study, Nanoscale, 2017, 9, 6315–6326 RSC.
- M. Marchioni, P.-H. Jouneau, M. Chevallet, I. Michaud-Soret and A. Deniaud, Silver nanoparticle fate in mammals: Bridging in vitro and in vivo studies, Coord. Chem. Rev., 2018, 364, 118–136 CrossRef CAS.
- K. Park, E.-J. Park, I. K. Chun, K. Choi, S. H. Lee, J. Yoon and B. C. Lee, Bioavailability and toxicokinetics of citrate-coated silver nanoparticles in rats, Arch. Pharmacal Res., 2011, 34, 153–158 CrossRef CAS PubMed.
- J. Bertinato, L. Cheung, R. Hoque and L. J. Plouffe, Ctr1 transports silver into mammalian cells, J. Trace Elem. Med. Biol., 2010, 24, 178–184 CrossRef CAS PubMed.
- A. Ibricevic, S. L. Brody, W. J. Youngs and C. L. Cannon, ATP7B detoxifies silver in ciliated airway epithelial cells, Toxicol. Appl. Pharmacol., 2010, 243, 315–322 CrossRef CAS PubMed.
- M. Linder, L. Wooten, P. Cerveza, S. Cotton, R. Shulze and N. Lomeli, Copper transport, Am. J. Clin. Nutr., 1998, 67, 965S–971S CrossRef CAS PubMed.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, What the Cell “Sees” in Bionanoscience, J. Am. Chem. Soc., 2010, 132, 5761–5768 CrossRef CAS PubMed.
- N. Durán, C. P. Silveira, M. Durán and D. S. T. Martinez, Silver nanoparticle protein corona and toxicity: a mini-review, J. Nanobiotechnol., 2015, 13(55) DOI:10.1186/s12951-015-0114-4.
- X. Jiang, T. Miclăuş, L. Wang, R. Foldbjerg, D. S. Sutherland, H. Autrup, C. Chen and C. Beer, Fast intracellular dissolution and persistent cellular uptake of silver nanoparticles in CHO-K1 cells: implication for cytotoxicity, Nanotoxicology, 2015, 9, 181–189 CrossRef CAS PubMed.
- L. Wang, T. Zhang, P. Li, W. Huang, J. Tang, P. Wang, J. Liu, Q. Yuan, R. Bai, B. Li, K. Zhang, Y. Zhao and C. Chen, Use of Synchrotron Radiation-Analytical Techniques To Reveal Chemical Origin of Silver-Nanoparticle Cytotoxicity, ACS Nano, 2015, 9, 6532–6547 CrossRef CAS PubMed.
- G. Veronesi, A. Deniaud, T. Gallon, P.-H. Jouneau, J. Villanova, P. Delangle, M. Carrière, I. Kieffer, P. Charbonnier, E. Mintz and I. Michaud-Soret, Visualization, quantification and coordination of Ag+ ions released from silver nanoparticles in hepatocytes, Nanoscale, 2016, 8, 17012–17021 RSC.
- G. Veronesi, C. Aude-Garcia, I. Kieffer, T. Gallon, P. Delangle, N. Herlin-Boime, T. Rabilloud and M. Carrière, Exposure-dependent Ag+ release from silver nanoparticles and its complexation in AgS2 sites in primary murine macrophages, Nanoscale, 2015, 7, 7323–7330 RSC.
- L. Bobyk, A. Tarantini, D. Beal, G. Veronesi, I. Kieffer, S. Motellier, E. Valsami-Jones, I. Lynch, P.-H. Jouneau, K. Pernet-Gallay, C. Aude-Garcia, S. Sauvaigo, T. Douki, T. Rabilloud and M. Carriere, Toxicity and chemical transformation of silver nanoparticles in A549 lung cells: dose-rate-dependent genotoxic impact, Environ. Sci.: Nano, 2021, 8, 806–821 RSC.
- V. Tardillo Suárez, E. Karepina, M. Chevallet, B. Gallet, C. Cottet-Rousselle, P. Charbonnier, C. Moriscot, I. Michaud-Soret, W. Bal, A. Fuchs, R. Tucoulou, P.-H. Jouneau, G. Veronesi and A. Deniaud, Nuclear translocation of silver ions and hepatocyte nuclear receptor impairment upon exposure to silver nanoparticles, Environ. Sci.: Nano, 2020, 7, 1373–1387 RSC.
- V. Tardillo Suárez, B. Gallet, M. Chevallet, P.-H. Jouneau, R. Tucoulou, G. Veronesi and A. Deniaud, Correlative transmission electron microscopy and high-resolution hard X-ray fluorescence microscopy of cell sections to measure trace element concentrations at the organelle level, J. Struct. Biol., 2021, 213, 107766 CrossRef PubMed.
- V. A. Solé, E. Papillon, M. Cotte, P. Walter and J. Susini, A multiplatform code for the analysis of energy-dispersive X-ray fluorescence spectra, Spectrochim. Acta, Part B, 2007, 62, 63–68 CrossRef.
- O. Proux, X. Biquard, E. Lahera, J. J. Menthonnex, A. Prat, O. Ulrich, Y. Soldo, P. Trvisson, G. Kapoujyan, G. Perroux, P. Taunier, D. Grand, P. Jeantet, M. Deleglise, J. Roux and J. Hazemann, FAME A New Beamline for XRay Absorption Investigations of VeryDiluted Systems of Environmental, Material and Biological Interests, Phys. Scr., 2005, 970 CrossRef CAS.
- O. Proux, V. Nassif, A. Prat, O. Ulrich, E. Lahera, X. Biquard, J.-J. Menthonnex and J.-L. Hazemann, Feedback system of a liquid-nitrogen-cooled double-crystal monochromator: design and performances, J. Synchrotron Radiat., 2006, 13, 59–68 CrossRef CAS PubMed.
- A. E. Pradas del Real, H. Castillo-Michel, R. Kaegi, B. Sinnet, V. Magnin, N. Findling, J. Villanova, M. Carrière, C. Santaella, A. Fernández-Martínez, C. Levard and G. Sarret, Fate of Ag-NPs in Sewage Sludge after Application on Agricultural Soils, Environ. Sci. Technol., 2016, 50, 1759–1768 CrossRef CAS PubMed.
- G. Veronesi, T. Gallon, A. Deniaud, B. Boff, C. Gateau, C. Lebrun, C. Vidaud, F. Rollin-Genetet, M. Carrière, I. Kieffer, E. Mintz, P. Delangle and I. Michaud-Soret, XAS Investigation of Silver(I) Coordination in Copper(I) Biological Binding Sites, Inorg. Chem., 2015, 54, 11688–11696 CrossRef CAS PubMed.
- I. Kieffer and D. Testemale, French Absorption spectroscopy beamline in Material and Environmental science, database service, 2016 Search PubMed.
- B. Ravel and M. Newville, ATHENA , ARTEMIS , HEPHAESTUS : data analysis for X-ray absorption spectroscopy using IFEFFIT, J. Synchrotron Radiat., 2005, 12, 537–541 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- P. Thevenaz, U. E. Ruttimann and M. Unser, Fiji: an open-source platform for biological-image analysis, IEEE Trans. Image Process., 1998, 7, 27–41 CrossRef CAS PubMed.
- D. G. Lowe, Distinctive Image Features from Scale-Invariant Keypoints, Int. J. Comput. Vis., 2004, 60, 91–110 CrossRef.
- D. Tschumperle and R. Deriche, Vector-valued image regularization with PDEs: a common framework for different applications, IEEE Trans. Pattern Anal. Mach. Intell., 2005, 27, 506–517 CAS.
- J. Fehrenbach, P. Weiss and C. Lorenzo, Variational Algorithms to Remove Stationary Noise: Applications to Microscopy Imaging, IEEE Trans. Image Process., 2012, 21, 4420–4430 Search PubMed.
- C. Sommer, C. Straehle, U. Kothe and F. A. Hamprecht, Ilastik: Interactive learning and segmentation toolkit, in 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, IEEE, Chicago, IL, USA, 2011, pp. 230–233 Search PubMed.
- I. Belevich, M. Joensuu, D. Kumar, H. Vihinen and E. Jokitalo, Microscopy Image Browser: A Platform for Segmentation and Analysis of Multidimensional Datasets, PLoS Biol., 2016, 14, e1002340 CrossRef PubMed.
- J. Ahrens, B. Geveci and C. Law, 36 - ParaView: An End-User Tool for Large-Data Visualization, in Visualization Handbook, ed. C. D. Hansen and C. R. Johnson, Butterworth-Heinemann, Burlington, 2005, pp. 717–731 Search PubMed.
- S. Bolte and F. P. CordelièRes, A guided tour into subcellular colocalization analysis in light microscopy, J. Microsc., 2006, 224, 213–232 CrossRef CAS PubMed.
- D. Legland, K. Kiêu and M.-F. Devaux, Computation of Minkowski Measures on 2D and 3D Binary Images, Image Anal. Stereol., 2011, 26, 83 CrossRef.
- A. Kermanizadeh, T. Berthing, E. Guzniczak, M. Wheeldon, G. Whyte, U. Vogel, W. Moritz and V. Stone, Assessment of nanomaterial-induced hepatotoxicity using a 3D human primary multi-cellular microtissue exposed repeatedly over 21 days - the suitability of the in vitro system as an in vivo surrogate, Part. Fibre Toxicol., 2019, 16(42) DOI:10.1186/s12989-019-0326-0.
- A. Espinosa, A. Curcio, S. Cabana, G. Radtke, M. Bugnet, J. Kolosnjaj-Tabi, C. Péchoux, C. Alvarez-Lorenzo, G. A. Botton, A. K. A. Silva, A. Abou-Hassan and C. Wilhelm, Intracellular Biodegradation of Ag Nanoparticles, Storage in Ferritin, and Protection by a Au Shell for Enhanced Photothermal Therapy, ACS Nano, 2018, 12, 6523–6535 CrossRef CAS PubMed.
- A. Curcio, A. V. de Walle, E. Benassai, A. Serrano, N. Luciani, N. Menguy, B. B. Manshian, A. Sargsian, S. Soenen, A. Espinosa, A. Abou-Hassan and C. Wilhelm, Massive Intracellular Remodeling of CuS Nanomaterials Produces Nontoxic Bioengineered Structures with Preserved Photothermal Potential, ACS Nano, 2021, 15, 9782–9795 CrossRef CAS PubMed.
- V. R. Sharma, A. Shrivastava, B. Gallet, E. Karepina, P. Charbonnier, M. Chevallet, P.-H. Jouneau and A. Deniaud, Canalicular domain structure and function in matrix-free hepatic spheroids, Biomater. Sci., 2020, 8, 485–496 RSC.
- K. Loeschner, N. Hadrup, K. Qvortrup, A. Larsen, X. Gao, U. Vogel, A. Mortensen, H. R. Lam and E. H. Larsen, Distribution of silver in rats following 28 days of repeated oral exposure to silver nanoparticles or silver acetate, Part. Fibre Toxicol., 2011, 8, 18 CrossRef CAS PubMed.
- M. van der Zande, R. J. Vandebriel, E. Van Doren, E. Kramer, Z. Herrera Rivera, C. S. Serrano-Rojero, E. R. Gremmer, J. Mast, R. J. B. Peters, P. C. H. Hollman, P. J. M. Hendriksen, H. J. P. Marvin, A. A. C. M. Peijnenburg and H. Bouwmeester, Distribution, Elimination, and Toxicity of Silver Nanoparticles and Silver Ions in Rats after 28-Day Oral Exposure, ACS Nano, 2012, 6, 7427–7442 CrossRef CAS PubMed.
- A. Malysheva, A. Ivask, C. L. Doolette, N. H. Voelcker and E. Lombi, Cellular binding, uptake and biotransformation of silver nanoparticles in human T lymphocytes, Nat. Nanotechnol., 2021, 16, 926–932 CrossRef CAS PubMed.
- K. Kluska, M. D. Peris-Díaz, D. Płonka, A. Moysa, M. Dadlez, A. Deniaud, W. Bal and A. Krężel, Formation of highly stable multinuclear Ag n S n clusters in zinc fingers disrupts their structure and function, Chem. Commun., 2020, 56, 1329–1332 RSC.
- K. Kluska, G. Veronesi, A. Deniaud, B. Hajdu, B. Gyurcsik, W. Bal and A. Krężel, Structures of Silver Fingers and a Pathway to Their Genotoxicity, Angew. Chem., Int. Ed., 2022 DOI:10.1002/anie.202116621.
- H. Zhang, X. Wang, M. Wang, L. Li, C. H. Chang, Z. Ji, T. Xia and A. E. Nel, Mammalian Cells Exhibit a Range of Sensitivities to Silver Nanoparticles that are Partially Explicable by Variations in Antioxidant Defense and Metallothionein Expression, Small, 2015, 11, 3797–3805 CrossRef CAS PubMed.
- D. A. Gonzalez-Carter, B. F. Leo, P. Ruenraroengsak, S. Chen, A. E. Goode, I. G. Theodorou, K. F. Chung, R. Carzaniga, M. S. Shaffer and D. T. Dexter, Silver nanoparticles reduce brain inflammation and related neurotoxicity through induction of H 2 S-synthesizing enzymes, Sci. Rep., 2017, 7, 42871 CrossRef PubMed.
- P. Charbonnier, B. Chovelon, C. Ravelet, T. D. Ngo, M. Chevallet and A. Deniaud, ATP7B-Deficient Hepatocytes Reveal the Importance of Protein Misfolding Induced at Low Copper Concentration, Cell, 2022, 11, 3400 CrossRef CAS PubMed.
- M. Minghetti and K. Schirmer, Interference of silver nanoparticles with essential metal homeostasis in a novel enterohepatic fish in vitro system, Environ. Sci.: Nano, 2019, 6, 1777–1790 RSC.
- J. Li, B. Zhang, X. Chang, J. Gan, W. Li, S. Niu, L. Kong, T. Wu, T. Zhang, M. Tang and Y. Xue, Silver nanoparticles modulate mitochondrial dynamics and biogenesis in HepG2 cells, Environ. Pollut., 2020, 256, 113430 CrossRef CAS PubMed.
- D. F. Mello, L. L. Maurer, I. T. Ryde, D. H. Songr, S. M. Marinakos, C. Jiang, M. R. Wiesner, H. Hsu-Kim and J. N. Meyer, In Vivo Effects of Silver Nanoparticles on Development, Behavior, and Mitochondrial Function are Altered by Genetic Defects in Mitochondrial Dynamics, Environ. Sci. Technol., 2022, 56, 1113–1124 CrossRef CAS PubMed.
- R. Holmila, H. Wu, J. Lee, A. W. Tsang, R. Singh and C. M. Furdui, Integrated Redox Proteomic Analysis Highlights New Mechanisms of Sensitivity to Silver Nanoparticles, Mol. Cell. Proteomics, 2021, 20, 100073 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3en00177f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |