Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Sticking and sliding of lipid bilayers on deformable substrates†
L.
Stubbington
a,
M.
Arroyo
b and
M.
Staykova
*a
aDurham University, UK. E-mail: margarita.staykova@durham.ac.uk
bUniversitat Politècnica de Catalunya, Barcelona, Spain
First published on 13th June 2016
Abstract
We examine here the properties of lipid bilayers coupled to deformable substrates. We show that by changing the extent of the substrate hydrophilicity, we can control the membrane–substrate coupling and the response of the bilayer to strain deformation. Our results demonstrate that lipid bilayers coupled to flexible substrates can easily accommodate large strains, form stable protrusions and open reversibly pores. These properties, which differ significantly from those of free standing membranes, can extend the applications of the current lipid technologies. Moreover, such systems better capture the mechanical architecture of the cell interface and can provide insights into the capacity of cells to reshape and respond to mechanical perturbations.
1. Introduction
Supported lipid bilayers (SLBs) have become a fundamental asset in the research on lipid and cell membranes. Among others, they have been used to assess the membrane structure and phase behavior,1 to reconstitute proteins, glycans and other cell membrane constituents,2 or to investigate cell membrane adhesion.3 In parallel to their wide biophysical applications, SLBs have become a promising platform for encapsulation, bio-sensing, separation and lipid nanotechnologies,4–9 albeit with modest technological success mainly due to their fragility. In this paper we discuss the emerging mechanical properties of lipid membranes supported on flexible substrates, and the novel biophysical and technological implications that these systems may provide.In current SLB systems, the role of the membrane support is mostly passive – it increases the mechanical stability of the membrane and facilitates the quantitative analysis of the membrane using a range of surface sensitive techniques.10 Its adverse effects on membrane fluidity and the structure of the reconstituted proteins are overcome by lifting the membrane from the solid support by means of polymer cushions, tethers or self assembled monolayers.2,10,11 In the last couple of years more and more studies have demonstrated that the support can also be used to actively manipulate the organization of the lipid membrane. In particular chemically heterogeneous or geometrically patterned substrates were shown to control the lateral organization of the lipid membrane.12–14
The requirement of a solid support has remained unchallenged probably due to the inability of freestanding lipid bilayers to sustain even modest stretch and compression.15 Intriguingly, nature has chosen to support its lipid structures onto elastic and actively reshaping polymeric networks, such as the actin cortex, the extracellular matrix and the basal lamina. Such architecture ensures that the cellular interface is malleable, responsive and yet robust. Moreover, as far as our current knowledge goes, mechanical stress imposed and transmitted through the cell membrane is a key regulator of cell physiology and differentiation, tissue morphogenesis, and embryogenesis.16 Creating artificial systems that capture the elasticity and deformability of the cell interface would provide invaluable insights into the mechanisms of mechano-transduction in cells, and the principles conferring biological membranes with the ability to dynamically remodel and to sustain mechanical stresses. We further expect that such systems will open new horizons for the current lipid technologies, including flexible biosensors and lipogel capsules that can reversibly change their shape, adhesivity and permeability in response to chemical and mechanical stimuli. The experiments reported here provide the background for such developments.
Previously we have demonstrated that a simple lipid bilayer coupled to an elastic polydimethylsiloxane (PDMS) substrate can follow the changes in the substrate area without losing its integrity.17,18 Upon substrate expansion, the bilayer absorbs lipid protrusions in order to increase its in-plane area; upon compression, it expels them back. The shape of the membrane protrusions can be controlled by the substrate strain and the osmotic pressure difference across the membrane.18 Our in vitro findings have been recently reproduced in cells coupled to elastic substrates,19 thus indicating that biomembranes also use purely physical mechanisms to accommodate fast changes in their surface area. In addition, the potential of using stimuli responsive gels coated with lipid shells for encapsulation and material release has shown promising results.20,21
In this paper we present further insights into the rich functionality of deformable supported lipid bilayers. We show that a simple modification of the surface properties of the elastic substrate by plasma oxidation can induce two very different mechanisms of remodeling in the lipid bilayer, which help buffer the applied stresses and preserve its integrity.
2. Methods and materials
2.1. Consumables
1,2-Dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Rh-DPPE) were purchased from Avanti Polar Lipids (Alabaster, AL) and used without further purification. Chloroform, trizma buffer, and sucrose were purchased from Sigma Aldrich. Polydimethylsiloxane (PDMS), curing agent (Sylgard 184 Silicone Elastomer Kit, catalog no. 240401 9862), microscope slides and cover glasses from VWR (catalog no. 48366 045) were used. For the preparation procedure of GUVs we used Indium Tin Oxide coated glasses (ITO glasses) from Delta Technologies (no. X180).2.2. Device
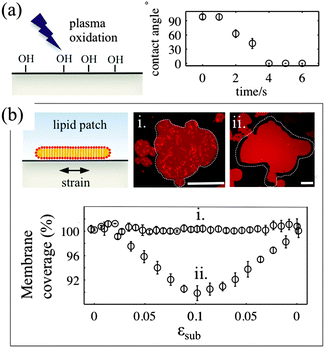
For the membrane strain experiments we use the same device as described previously by us.17 A thin circular sheet of cured PDMS 1–1.5 mm in diameter is suspended above the outlet of a microfluidic channel. The channel inlet is connected to a machine driven syringe pump (Harvard PhD apparatus). Application of a positive pressure to the syringe causes the thin sheet of PDMS to expand from a flat geometry to a hemispherical cap, the center of which is subject to a biaxial area expansion. Lipids coupled to this substrate are thus required to respond to this change in area. A typical experiment involves the inflation of the device up to a total substrate area change of between 12 and 20% and subsequent deflation, with strain rates between 0.001 and 0.8% s−1. The nominal strain rate is 0.05% s−1.The PDMS surface onto which the membrane is deposited is exposed to a low-pressure air plasma (VacuLAB Plasma Treater, Tantec) for a duration of 0–30 seconds. The substrate hydrophilicity is assessed by measuring the contact angle of a 10 μl aqueous droplet deposited on the surface immediately after treatment.
2.3. Lipid patches
Immediately after exposure to plasma the device is wetted with TRIS buffer (13 mM Trizma base, 150 mM NaCl, 2 mM CaCl2). A small amount of solution of giant unilamellar vesicles (GUVs) is added to the incubation chamber and the device is left, covered, for 10 minutes during which time GUVs sediment to the bottom of the chamber and eventually rupture on the substrate surface, forming a fluid and continuous lipid bilayer patch. For the experiments, the chamber is gently washed with TRIS buffer or with water to remove unfused vesicles.GUVs, composed of DOPC and Rh-DPPE in 99.5/0.5 molar ratio, are formed by standard electro-formation techniques described elsewhere17 and are kept for a maximum of 2 days prior to experiments.
2.4. Imaging
The imaging of the membrane response to substrate deformation is done using an inverted optical microscope Nikon Eclipse Ti-E and recorded using an ANDOR camera Neo 5.5 sCMOS (Oxford Instruments). The integrated perfect focusing system (PFS) in the microscope allows us to follow automatically the PDMS surface which changes its focal plane during the strain deformation (inflation and deflation). FRAP experiments are carried out using an inverted Nikon confocal microscope.2.5. Image analysis
Image analysis is performed using ImageJ. During stress–strain experiments, the bright-field is sampled at a frequency one third of the frame rate of the fluorescence images. From the sequence of bright-field images one can assess the area change of the substrate by tracking the displacement of small defects (air bubbles) in the PDMS, which become visible under bright-field illumination.The changes in bilayer area in response to this substrate stress are determined by first background subtracting the fluorescence images and then applying an appropriate threshold to generate a binary stack. Particle analysis can then be used to track the area of the patch over time by filtering results according to size. If the bilayer opens pores during the expansion a secondary binary image stack is generated to measure the total pore area in each frame. This is subsequently subtracted from the total membrane area. Data acquired from ImageJ is transferred to MATLAB for subsequent analysis, fits and plotting.
2.6. Contact angle measurements
To measure the static and dynamic contact angles of 10 μl aqueous droplets on plasma oxidised PDMS substrates as a function of the plasma exposure time we use a previously described experimental setup22 and an Image J plug-in.3. Results
3.1. Substrate surface properties determine the mechanisms of stress relaxation in lipid bilayers
The deformable PDMS substrates are exposed to plasma oxidation for various time durations. As shown previously, plasma oxygen hydroxylates the surface by increasing the number of silanol groups (–Si–OH), and increases its hydrophilicity.23 This is confirmed by the larger contact angle that aqueous droplets form on shortly oxidized PDMS substrates. For plasma exposures longer than 4–5 seconds we observe complete wetting of the PDMS substrate by the droplet (Fig. 1a). To characterise the effects of plasma exposure on the PDMS surface properties on the microscale we have measured the dynamic contact angles of advancing and receding droplets on the PDMS substrate upon increasing and decreasing their volumes, respectively (Fig. S1, ESI†). The relatively large contact angle hysteresis, which decreases from 50° to 35° as the plasma exposure time increases, indicates that there are chemical and/or mechanical surface heterogeneities24 that are more pronounced for short exposure times.In our experiments, we use two types of substrates to support the membrane: (1) partly hydrophilic PDMS (3 seconds of plasma exposure), on which the droplet contact angle is between 35° and 60°, and (2) hydrophilic PDMS (30 seconds plasma oxidation), which appears completely wetted by an aqueous droplet. The membrane patches are formed by GUV fusion on freshly oxidized substrates and left to equilibrate for 20–30 min before the strain application (Fig. 1b). All membrane patches appear unilamellar, as indicated by our AFM measurements (Fig. S2, ESI†), as well as by their homogeneous fluorescence appearance, except for the brighter lipid protrusions on top of the membranes. Note that PDMS substrates that have been exposed for less than 2 seconds to plasma do not support the formation of lipid bilayer patches. In line with previous observations23 we observe the formation of lipid monolayers with half of the fluorescence intensity of the bilayer membrane (data not shown).
For the membrane strain experiments we usually apply a cycle of substrate biaxial expansion, followed by compression, and image the response of the fluorescent bilayer patch by optical microscopy. By using discontinuous patches where we can image both the planar part of the bilayer and the patch perimeter we can quantify changes in the shape and the surface area of the membrane as a function of the substrate strain. Our results show that depending on the substrate hydrophilicity, lipid bilayers exhibit different mechanisms to sustain the substrate deformation without rupturing (Fig. 1b).
On partly hydrophilic substrates, the membrane is pinned to the substrate and follows closely the changes in the substrate area (“sticky” membranes). Consequently, the substrate remains completely covered by the membrane throughout the whole strain cycle (Fig. 1b–i). To sustain the in-plane area changes uncompromised, the membrane releases or acquires extra lipids through out-of plane lipid protrusions, similar to what we have previously shown.17
On hydrophilic surfaces, the behaviour of the lipid membrane differs substantially. When altering the area of the substrate, the membrane decouples from it and avoids the imposed dilation by sliding over the deforming substrate (hence “sliding” membranes). As a result the membrane surface coverage decreases upon substrate expansion, and increases back, upon consequent compression (Fig. 1b–ii).
We will next discuss the mechanisms of the sticky and sliding membrane behaviour separately, and under what conditions they cannot be sustained anymore and the membrane forms pores upon stretching.
3.2. Sticky membranes
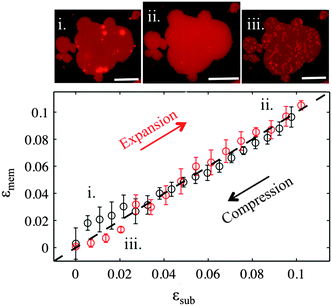
PDMS substrates exposed to plasma for 3 seconds cannot induce fusion of small unilamellar vesicles, in accordance with previous studies.23 However larger GUVs readily fuse due to their large size and form continuous bilayer patches, as confirmed by FRAP experiments (Fig. S3, ESI†). The patches usually exhibit numerous multilamellar spherical protrusions on top, which get absorbed into the planar membrane, as it matches the substrate expansion (Video S1, ESI†). When the membrane is compressed back to its initial dimensions the excess lipids are expelled as tubular protrusions, which remain stable for at least 30 minutes after the end of the compression (Fig. S4 and Video S2, ESI†). The change of the shape in the protrusions from spherical to tubular is associated with the loss of volume during the initial fusion process and presumably to the reorganization from multilamellar structures into unilamellar tubules.18Upon large substrate expansion, usually εsub of about 10%, the available reservoir of lipid protrusions becomes insufficient to sustain the membrane area increase and the membrane patch ruptures (Fig. S5, ESI†). Note that in our previous experiments, continuous supported bilayers were easily expanded by up to 30–40% before rupture due to their large amount of reservoir.17,18 Interestingly the ruptured patches are also able to form tubes upon compression suggesting that even in this state lipids are able to flow (Fig. S5, ESI†). Indeed, we observe fluorescence recovery after photobleaching in all sticky membrane patches, albeit slower in the ruptured patches where the lipids need to travel along a network of pores (Fig. S3, ESI†).
3.3. Sliding membranes
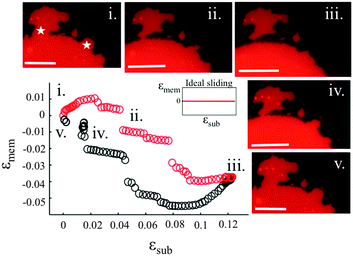
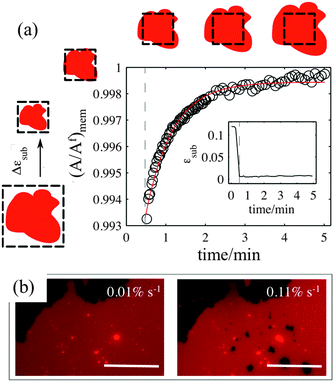
Our experiments show that on hydrophilic PDMS substrates (oxidized for 30 seconds) the membrane patch does not follow the deformation of the substrate but instead slides over it (Video S3 and S4, ESI†). In an ideal sliding case, i.e. if the membrane were completely decoupled from the substrate, its surface area would remain constant upon substrate deformation (Fig. 3(inset)). Our measurements however show that the membrane exhibits a complex area relaxation behaviour. Up to approximately 2% strains, the patch expands simultaneously with the substrate (Fig. 3). When a critical applied strain is reached, the membrane starts sliding, by retracting lipids from the periphery towards the center of the patch. During these lipid flows, the patch leaves the disconnected lipid island behind. Because we define strain through the area of the continuously connected patch the detachment of these islands accounts for the small step-wise area decrease in the sliding patch (Fig. 3ii and iii). Similarly, upon substrate compression the membrane initially follows the substrate before it sets into outward sliding. This stick-slip behavior upon extension and compression suggests the existence of a yield interfacial traction between the bilayer and the substrate, below which the membrane is pinned and above which it slides. As lipids flow against the shrinking substrate they follow the same path in which they retracted. The patch gains lipid area by merging with the previously disconnected lipid islands (Fig. 3iv). During the strain cycle the patch significantly changes its shape upon expansion (Fig. 3i and iii) and restores it upon compression (Fig. 3iv).3.4. Pore formation
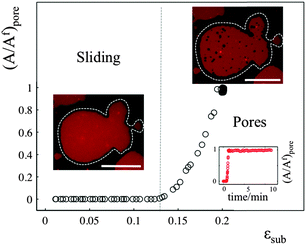
The sliding mechanisms cannot indefinitely accommodate substrate deformation, and beyond a certain strain (about 10% in the experiment reported in Fig. 4) most of our patches rapidly form pores. While the pores grow as the substrate is expanded, they also change their shape, implying that there is a lipid flow away from the pore to the continuous part of the bilayer (Video S5, ESI†). At the same time, the sliding of the patch perimeter ceases. Pores in supported bilayers are stable as long as the substrate underneath is kept at constant strain (Fig. 4), suggesting that the line tension acting on the periphery of the pores is not sufficient to overcome the yield traction required for sliding. Upon substrate compression the pores reseal first, before the outward sliding of the contour begins. Interestingly, pore opening and resealing during extension–compression cycles exhibit some amount of hysteresis (Fig. S6, ESI†), consistent with the notion of a yield interfacial traction required for sliding.The opening of pores in these membranes resting on a hydrophilic substrate suggests that the sliding mechanism cannot fully relax the dilational stress imposed on the membrane. One possible explanation would be the existence of frictional tractions at the membrane–substrate interface dynamically opposing sliding, which would build up tension in the bilayer. To test this hypothesis, we subject patches to a rapid expansion or compression and image the area of the patch after the end of the strain pulse. Fig. 5a shows that the membrane area continues to increase for some time after the end of the fast compression. The area change can be fitted to an exponential with a time constant of about τ = 40 ± 3 seconds. Similar values are obtained from the rapid expansion experiments on the same patch. Experiments on different samples show relaxation times varying between 15 and 50 seconds (Table S1, ESI†). The exponential evolution towards equilibrium suggests that the nature of the interaction with the substrate dynamically opposing sliding is a viscous friction, by which the force per unit area (traction) opposing sliding is proportional to the sliding velocity. To rationalize these measurements, we assume that the dominant effect driving area changes is bilayer compressibility, characterized by the compressibility modulus K ∼ 0.1 N m−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 and that the dominant dissipative effect is given by the viscous friction coefficient bs. From elementary dimensional analysis considerations, the relaxation time is given by τ ∼ bsA/K, where A is the area of the patch. In our example, A ∼ 12
25 and that the dominant dissipative effect is given by the viscous friction coefficient bs. From elementary dimensional analysis considerations, the relaxation time is given by τ ∼ bsA/K, where A is the area of the patch. In our example, A ∼ 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 850 μm2 resulting in bs ∼ 3 × 108 N s m−3, which agrees with the reported values in the literature for membranes on hydrophilic substrates probed using hydrodynamic shear forces.26,27
850 μm2 resulting in bs ∼ 3 × 108 N s m−3, which agrees with the reported values in the literature for membranes on hydrophilic substrates probed using hydrodynamic shear forces.26,27
If membrane sliding is opposed by an interfacial viscous friction, then it should be possible to prevent the opening of pores by reducing the rate of substrate expansion. In agreement with this prediction, large patches (where relaxation time should be longer) stretched at a strain rate of 0.11% s−1 show pores after 10% expansion, whereas the same samples expanded to the same strain amplitude at 0.01% s−1 remain continuous (Fig. 5b and Fig. S7, ESI†).
4. Discussion
Our results demonstrate that the response of lipid membranes to substrate strain deformation depends strongly on the hydrophilicity of the substrate. On partly hydrophilic surfaces the membrane–substrate coupling is so strong that the bilayer appears to be glued to the substrate underneath and is forced to change its area simultaneously with it. A deformation in the substrate area results in changes of the membrane lipid density, which are compensated by the absorption and expulsion of out-of-plane lipid protrusions (Fig. 2). These findings are very similar to our previous results on continuous supported bilayers upon strain deformation, which we have justified by the lateral confinement of the lipids and the interstitial liquid (between the substrate and the bilayer).17,18 The present study demonstrates that in the absence of such lateral constraints the strong coupling of the membrane to the substrate deformation is sufficient to trigger out-of plane lipid remodeling.The response of the sticky membrane to substrate area changes portrays a seeming contradiction. On the one hand, the formation of lipid tubes upon compression implies that lipids can move freely over the substrate and flow into the extending out-of-plane protrusions. On the other hand, the long range collective motion of lipids relative to the substrate appears completely inhibited. This is experimentally evident from (1) the absence of changes in the patch contour during the strain cycle (Fig. 2); (2) the longevity of the projected lipid tubes (Fig. S4, ESI†); and (3) the fact that ruptured patches prefer to form tubes upon subsequent compression than to close their pores (Fig. S5, ESI†). These observations can be reconciled by our contact angle measurements, which suggest that PDMS substrates following short plasma exposures are chemically and/or mechanically heterogeneous (Fig. S1, ESI†). Such heterogeneities will likely influence the organization or the lipids and their interaction with the substrate, and may function as pinning sites for the membrane. Discretely pinned membranes would allow the molecular motion of lipids and maybe the collective flow on small scales (necessary for the formation of protrusions) but not a coherent large-scale lipid motion past the substrate.
Lipid membranes supported on hydrophilic substrates exhibit a very different behaviour. The lipids slide over large scales to allow the membrane to preserve its area upon area changes in the underlying substrate (Fig. 3). Furthermore, we have never observed the absorption of lipid protrusions upon stretch even though they are readily available in these patches as well. Such behaviour suggests that the coupling of the membrane to hydrophilic substrates is weaker than the cohesive interaction between the lipids, and sliding is preferable to membrane deformation.
Our relaxation experiments indicate that the membrane–substrate coupling exhibits a complex rheology. On hydrophilic surfaces, bilayers need to overcome a critical yield interfacial traction to begin sliding, and then sliding is dynamically opposed by a viscous friction. Previous studies suggest that this behavior could be related to the lubrication properties of the water hydration layer between the membrane and the substrate.28,29 Substrates treated longer with plasma exhibit a higher density of uniformly distributed –Si–OH groups that favor the formation of a stable hydration layer, where not all water molecules are ordered and can be sheared upon stress application.28,30,31 In contrast, on shortly oxidized substrates, the low and heterogeneous density of hydroxyl groups leads to a much thinner and highly structured water layer,30 which strongly hinders the relative motion of lipids past the substrate.
Finally, we should mention that since friction force scales with the surface area, the observed membrane behaviour on deformable substrates is very likely to depend on the size of the membrane patch. Thus, both the relaxation time of the membrane and the critical strain at which sliding occurs are expected to increase for larger patches. This may explain why in our previous experiments with continuous supported bilayers we were able to observe only the stick behaviour.17,18 However, our current experiments with discrete membrane patches failed to verify the size dependence (Tables S1 and S2, ESI†), likely due to the small variabilty in the patch sizes and our inability to precisely control the PDMS surface properties using our plasma device.
5. Conclusions
In summary, we have shown in this paper that supported lipid membranes can adapt to changes in the substrate area by recruiting out-of plane lipid reservoirs or by sliding. The nature of the response is determined by the extent of hydrophilicity of the supporting surface. The possibility of (1) opening and closing membrane pores on demand, (2) making planar membranes to expel diverse protrusions, and (3) controlling the area of lipid coverage of the substrate can lay the foundations of exciting new technological developments. In the future, we will aim to develop more precise methods to control the substrate interfacial properties, and to translate our 2D findings in the design of mechano-responsive capsules for controlled delivery. Moreover, in order to obtain further insights into the links between the mechanical architecture and the functionality of the cell interface we will develop systems that use biologically relevant substrates and include different modes of membrane–substrate binding, including molecular linkers.Acknowledgements
MS and LS acknowledge the financial support of Durham University, the Biophysical Sciences Institute (BSI), and EPSRC-UK. We thank Dr Kislon Voitchovsky for useful discussions, and E. Miller, M. Masukawa, and Prof. J. Girkin for experimental support.Notes and references
- L. K. Tamm and H. M. McConnell, Biophys. J., 1985, 47, 105 CrossRef CAS PubMed.
- M. Tanaka and E. Sackmann, Nature, 2005, 437, 656 CrossRef CAS PubMed.
- J. T. Groves and M. L. Dustin, J. Immunol. Methods, 2003, 278, 19 CrossRef CAS PubMed.
- C. E. Ashley, et al. , Nat. Mater., 2011, 10, 389 CrossRef CAS PubMed.
- E. Reimhult and K. Kumar, Trends Biotechnol., 2008, 26, 82 CrossRef CAS PubMed.
- C. H. Nielsen, Anal. Bioanal. Chem., 2009, 395, 697 CrossRef CAS PubMed.
- S. Mashaghi, T. Jadidi, G. Koenderink and A. Mashaghi, Int. J. Mol. Sci., 2013, 14, 4242 CrossRef CAS PubMed.
- A. Wagner and K. Vorauer-Uhl, J. Drug Delivery, 2011, 2011, 591325 Search PubMed.
- G. Bozzuto and A. Molinari, Int. J. Nanomed., 2015, 10, 975 CrossRef CAS PubMed.
- J. A. Jackman, W. Knoll and N. J. Cho, Materials, 2012, 5, 2637 CrossRef CAS.
- M. L. Wagner and L. K. Tamm, Biophys. J., 2000, 79, 1400 CrossRef CAS PubMed.
- A. N. Parikh, Biointerphases, 2008, 3, FA22 CrossRef CAS PubMed.
- A. B. Subramaniam, G. Guidotti, V. Manoharan and H. A. Stone, Nat. Mater., 2013, 12, 128 CrossRef CAS PubMed.
- J. T. Groves and S. G. Boxer, Acc. Chem. Res., 2002, 35, 149 CrossRef CAS PubMed.
- D. Needham and R. S. Nunn, Biophys. J., 1990, 58, 997 CrossRef CAS PubMed.
- T. Wyatt and B. Baumand G. Charras, Curr. Opin. Cell Biol., 2016, 38, 68 CrossRef CAS PubMed.
- M. Staykova, D. P. Holmes, C. Read and H. A. Stone, PNAS, 2011, 108, 9084 CrossRef CAS PubMed.
- M. Staykova, M. Arroyo, M. Rahimi and H. A. Stone, Phys. Rev. Lett., 2013, 110, 1 CrossRef PubMed.
- A. J. Kosmalska, et al. , Nat. Commun., 2015, 6, 7292 CrossRef CAS PubMed.
- Q. Saleem, B. Liu, C. C. Gradinaru and P. M. Macdonald, Biomacromolecules, 2011, 12, 2364 CrossRef CAS PubMed.
- Y. Wang, S. Tu, A. N. Pinchuk and M. P. Xiong, J. Colloid Interface Sci., 2013, 406, 247 CrossRef CAS PubMed.
- G. Lamour, et al. , J. Chem. Educ., 2010, 87, 1403 CrossRef CAS.
- P. Lenz, C. M. Ajo-Franklin and S. G. Boxer, Langmuir, 2004, 20, 11092 CrossRef CAS PubMed.
- J. N. Israelachvili, Intermolecular and Surface Forces, Intermolecular and Surface Forces, Elsevier, 2011, pp. 415–467 Search PubMed.
- R. Dimova, et al. , J. Phys.: Condens. Matter, 2006, 18, 1151 CrossRef PubMed.
- P. Jönsson and F. Höök, Langmuir, 2011, 27, 1430 CrossRef PubMed.
- P. Jönsson, J. P. Beech, J. O. Tegenfeldt and F. Höök, J. Am. Chem. Soc., 2009, 131, 5294 CrossRef PubMed.
- J. Klein, Friction, 2013, 1, 1 CrossRef CAS.
- R. Merkel, E. Sackmann and E. Evans, J. Phys., 1989, 50, 1535 CAS.
- S. Ahmed, R. R. Madathingal, S. L. Wunder, Y. Chen and G. Bothun, Soft Matter, 2011, 7, 1936 RSC.
- R. Tero, Materials (Basel), Multidisciplinary Digital Publishing Institute, 2012, vol. 5, p. 2658 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6sm00786d |
| This journal is © The Royal Society of Chemistry 2017 |