Open Access Article
Open Access ArticleIntegrated assessment of the elimination of particle-associated fecal indicators in algal–bacterial granule photobioreactors†
Waleed M. M.
El-Sayed
 ab,
Tengge
Zhang
ab,
Tengge
Zhang
 a,
Matthew E.
Verbyla
c and
Meng
Wang
*a
a,
Matthew E.
Verbyla
c and
Meng
Wang
*a
aDepartment of Energy and Mineral Engineering and EMS Energy Institute, Pennsylvania State University, University Park, PA 16802, USA. E-mail: mxw1118@psu.edu
bMarine Microbiology Lab., National Institute of Oceanography and Fisheries, Cairo, Egypt
cDepartment of Civil, Construction, and Environmental Engineering, San Diego State University, San Diego, California 92182, USA
First published on 11th March 2025
Abstract
Two photo-sequencing batch reactors (PSBR) fed with real wastewater were evaluated to understand the elimination and particle association of fecal indicator bacteria (FIB) and coliphages. The average log10 removal of E. coli and Enterococcus spp. were 3.2 and 2.9, respectively, for the PSBR with a low airflow rate of 0.2 LPM (PSBR-L), and 2.8 and 2.7, respectively, for the PSBR with a high airflow rate of 0.5 LPM (PSBR-H). The average log10 removals of F-specific and somatic coliphages were 2.9 and 3.2, respectively, for the PSBR-L reactor, and 2.5 and 3.1, respectively, for the PSBR-H reactor. FIB had a maximum association on the 20–0.45 μm particles (46.1–63.3%), while the coliphages had the highest association on the 0.45–0.03 μm particles (44.7–51.2%). The dynamic adaptations in the microbial community structure (16S rRNA gene) were also investigated during the operation period. Genera involved in nutrient removal, such as Thauer spp., and Nitrospira spp., were detected across samples. The outcomes reveal the efficiency of photobioreactors in removing pathogen indicators from real wastewater.
Water impactThe contamination of water sources with fecal indicators is a significant public health concern, as these organisms indicate the presence of pathogens in water. Algal–bacterial systems offer a sustainable approach for removing fecal indicator bacteria (FIB) and coliphages from wastewater. This study investigates the particle association patterns of these indicators and identifies nutrient-removing bacteria, such as Thauer spp. and Nitrospira spp., that enhance the ecological benefits of this technology. The research contributes to sustainable water treatment solutions and aligns with the global objective of improving water quality through innovative photobioreactor technology. We anticipate that our findings, published in Environmental Science: Water Research & Technology, will inspire further research and practical applications in sustainable water treatment. |
1. Introduction
Algal–bacterial granular systems have shown promise as an innovative approach for biological wastewater treatment.1 The synergistic effects of algae and bacteria in these systems can improve the treatment processes by enhancing the settleability of the granules and removing contaminants such as nitrogen, chemical oxygen demand (COD), heavy metals, and pathogens from wastewater.2 Through promoting algal–bacterial aggregation and forming dense granules, biomass can be separated from treated water via sedimentation.3,4 Despite the effectiveness of this approach in terms of contaminant removal efficiency, the efficiency of algal–bacterial granules in pathogen removal has yet to be fully explored.The World Health Organization (WHO) guidelines recognize the critical impact that pathogenic microorganisms have in the transmission of disease via water resources and agricultural reuse. Enterococcus spp. and Escherichia coli are commonly used as indicators of fecal contamination in water resources, as well as in agricultural and aquaculture water reuse.5 However, they may not be adequate indicators of the presence of enteric viruses, which are more tolerant to treatment processes than bacteria.6 Coliphages, specifically somatic and F-specific coliphages, have been suggested as appropriate indicators of the microbiological quality of treated wastewater.6 Somatic coliphages are a diverse group of bacteriophages that can infect E. coli and other coliform bacteria, making them the most abundant group of indicator bacteriophages in environmental samples.7 F-specific coliphages, on the other hand, can infect E. coli and other coliform bacteria through the transferable F-plasmid, which is encoded in the sexual pili and can be transferred to enteric bacteria via conjugation.8 These coliphages are considered to be more suitable indicators of fecal pollution in bodies of water and may provide a more accurate and reliable means of evaluating the safety of water resources for reuse in agriculture and aquaculture.9
Previous studies have focused on the growth of microalgae in photobioreactors and biofilm reactors for disinfection purposes, but little is known about the removal of coliphages in these systems.10,11 Most studies of wastewater pathogens only consider the concentrations of pathogens in the fluid, and thus underestimate the levels of pathogens in the biomass, which can be highly contaminated.12 Wastewater treatment processes generate biomass that is often reused for land application.13,14 However, there is a knowledge gap regarding the particle association behavior of potential pathogens. Although several studies have characterized particles in wastewater, the nature of the associations between pathogens and particles remains unclear.15,16 In particular, little is known about the particle association with algal–bacterial granules, whose compact structure and biofilm layers provide an ideal environment for pathogens to attach and become part of the ecosystem. Elucidating the partitioning of pathogens is essential to facilitate system optimization to improve the efficiency, stability, and effectiveness of pathogen removal in granular reactors.
In this study, two photobioreactors with different airflow rates were operated to facilitate the formation of algal–bacterial granules for wastewater treatment. The specific objectives of this research are (1) to assess the impact of different airflow rates on the removal of fecal indicator bacteria (E. coli and Enterococcus spp.) and coliphages; (2) to understand the association of these microbial indicators with particles of various sizes; (3) to reveal the relationship between airflow rates, reactive oxygen species (ROS) generation and indicator bacteria and coliphage removal in wastewater treatment photobioreactors; and (4) to elucidate the impact of airflow rates on the migration of the microbial community compositions.
2. Materials and methods
2.1. Reactor operation and sample collection
The design and operation of the algal–bacterial granular photobioreactors was detailed by Zhang et al.17 Briefly, two laboratory-scale photo-sequencing batch reactors (PSBRs) with a 2.5 L working volume were operated at room temperature (25 ± 2 °C). The airflow rates for PSBR-L and PSBR-H were set at 0.2 L min−1 and 0.5 L min−1, respectively. The PSBRs were fed with real wastewater collected from the Penn State Water Reclamation Facility (WRF) every week and stored in a refrigerator at 4 °C for further feed. The NH4+ concentrations of the influent were measured using an ion chromatography (IC) system (DIONEX AQUINO, Thermo Scientific). The COD was determined using Orbeco-Hellige mid-range (0–1500 mg L−1) COD kits following standard method 5200B, while the total suspended solids (TSS) and volatile suspended solids (VSS) were measured according to standard methods.18 The influent wastewater had average TSS and VSS concentrations of 59.5 ± 34.5 mg L−1, and 39.4 ± 15.1 mg L−1, respectively. The average COD and NH4+-N concentrations were 155.6 ± 48.8 mg L−1 and 52.4 ± 18.2 mg L−1, respectively. Three stages of operations (set-up stage, stage 1, and stage 2) were implemented with varying light phases and feeding conditions. FIB and viruses were monitored during stage 2 (day 75–day 198), when the reactors achieved complete denitrification. Influent and effluent samples were collected weekly for FIB and virus analysis. 18 samples were collected for FIB analysis and 9 for virus analysis. All samples were stored at 4 °C and analyzed within 24 hours of collection. Each measurement was conducted in triplicate.2.2. Quantification of fecal indicator bacteria and coliphages
The quantification of E. coli and Enterococcus spp. was conducted using membrane filtration based on Environmental Protection Agency (EPA) Methods 1103.1 and 1600, respectively. Coliphages were evaluated using a double agar plaque assay based on the protocols described in Standard Methods 9224B and 9224C. The host bacteria E. coli Famp (ATCC#700891) and E. coli CN13 (ATCC#700609) were used for F-specific and somatic coliphages, respectively. The details of the quantification of FIB and coliphages are provided in the ESI.†The log10 removal of the four indicator groups was calculated from the difference between their concentrations in the wastewater influent and the effluent according to eqn (1).
 | (1) |
2.3. Particle association and size fraction of the fecal indicator bacteria and coliphages in the photobioreactor
Samples collected from PSBRs were filtered via cascade filtration for the particle association study (Fig. S1†).20,21 Briefly, 10 mL of each sample was sequentially filtered through 180 μm and 20 μm nylon net filters (Millipore) and 0.45 μm and 0.03 μm polyethersulfone membrane filters (Sterlitech). The qualified influence of each size portion was obtained by dividing the FIB and coliphage concentrations on each filter by the totality of the concentrations on all the filters for that sample.2.4. Detection of reactive oxygen species
The presence of ROS was detected using the K936-100-250 ROS detection assay kit (BioVision) according to the manufacturer's protocol. The method is briefly described in the supplementary material, and the results using fluorescence microscopy are illustrated in Fig. S2 and S3.† Additionally, the quantification of the ROS “superoxide” was determined by adding 0.5 mL of acetonitrile to a cuvette containing a wastewater sample. The mixture's absorbance was determined using a UV spectrophotometer at a wavelength of 255 nm. The molar concentration of superoxide was determined using Beer's law22 according to eqn (2): | (2) |
2.5. Impact of wastewater on reactive oxygen species production
Batch experiments of ROS production by algal–bacterial consortia in real and synthetic wastewater systems were set up to evaluate the kinetics of ROS production. Algal–bacterial consortia collected from a stock culture (originally obtained from the settling tank of the Penn State WRF and grown in BG11 medium for over a year), which was also the inoculum for the PSBRs, were inoculated in real wastewater and synthetic wastewater under light with an intensity of 200 ± 15 μmol m−2 s−1 and dark conditions on a shaking table at 120 rpm for 48 h. The synthetic wastewater composition is detailed in the ESI.† Superoxide was monitored as an indicator of ROS levels, and the data was collected in triplicate. The kinetics of the ROS production were evaluated using a first-order reaction model (eqn (3)) and the Gompertz model (eqn (4)) implemented by Matlab (R2023b).| [A] = a − be−k1t | (3) |
[A] = A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−exp(B − k2t)) exp(−exp(B − k2t)) | (4) |
2.6. Microbial community analysis
Biomass samples from both bioreactors, and influent and effluent samples were collected on day 189 for 16S rRNA analysis. The method for DNA extraction and sequencing is described in the ESI.† Alpha diversity indices, including Alpha Shannon, Simpson, Chao1, and ACE, were used to estimate within-species diversity.23 The “Venn diagram” package was used to visualize the relationships among distinct samples of the microbial community at the genus level (Chen and Boutros, 2011). The Silva 138.1 database was used as a reference database for taxonomic classification. All sequence data was submitted to the Sequence Read Archive (SRA) database at the NCBI under the accession number PRJNA1100726.2.7. Data analysis
The obtained data were compared via t-test using Microsoft Office Excel 2019, or one-way analysis of variance (ANOVA) at an alpha statistical significance level of 0.05 using Minitab statistical software. Using SPSS 26.0, the correlation coefficient between ROS production and pathogen indicators in mixed liquor samples was checked using Spearman's rank correlation coefficient (r > 0.5, p < 0.05).3. Result and discussion
3.1. Removal of fecal indicator bacteria and coliphages in the PSBRs
The effects of airflow rates on photobioreactor performance and microbial communities were reported in Zhang et al.17 In summary, higher air flow rates enhanced the nitrification rate and promoted the formation of compact, stable granules by increasing shear forces within the reactor. A nitrification rate of 4.3 mg N L−1 h−1 was achieved for PSBR-H (0.5 LPM), which was associated with an increase in the relative abundance of Nitrospira spp. and a decrease in the relative abundances of cyanobacteria and Chlorella spp. The average particle sizes for PSBR-L and PSBR-H were 0.29 ± 0.07 mm and 0.25 ± 0.1 mm, respectively. However, no significant differences were observed (p > 0.05).The coliphage and FIB concentrations in the raw wastewater (influent) remained constant over the experimental period (day 75–day 198) (Table 1). In the influent, E. coli had the highest average concentration, followed by Enterococcus spp. and then F-specific and somatic coliphages (Table 1). The concentrations of coliphages in raw wastewater have been observed to be in the range of 1.00 × 104 to 1.00 × 106 PFU per 100 mL.24–26 The concentration of E. coli and Enterococcus spp. in raw wastewater ranges from 1.00 × 104 to 1.00 × 109 CFU per 100 mL,16,27,28 depending on the evaluation method, geographic location, population, and wastewater characteristics.
| Stage | Pathogen indicator | Microbial concentrations CFU/100 ml or PFU/100 ml | log10 removal | N | ||
|---|---|---|---|---|---|---|
| Mean | Maximum | Minimum | ||||
| N = number of measurements. Means reported are the geometric means of the concentrations (mean ± SD). | ||||||
| Influent (raw wastewater) | Escherichia coli | 5.10 × 107 | 4.61 × 108 | 8.10 × 106 | — | 18 |
| Enterococcus spp. | 1.36 × 107 | 3.63 × 107 | 2.75 × 106 | — | 18 | |
| F-specific coliphages | 1.11 × 107 | 2.06 × 107 | 4.35 × 105 | — | 9 | |
| Somatic coliphages | 1.05 × 107 | 1.82 × 107 | 5.56 × 106 | — | 9 | |
| Effluent PSBR-L | Escherichia coli | 9.46 × 104 | 5.50 × 105 | 2.50 × 102 | 3.2 ± 0.39 | 18 |
| Enterococcus spp. | 9.62 × 104 | 5.20 × 105 | 1.00 × 103 | 2.9 ± 0.19 | 18 | |
| F-specific coliphages | 2.01 × 104 | 1.05 × 105 | 3.67 × 103 | 2.9 ± 0.19 | 9 | |
| Somatic coliphages | 4.8 × 103 | 9.50 × 103 | 2.00 × 103 | 3.4 ± 0.07 | 9 | |
| Effluent PSBR-H | Escherichia coli | 1.82 × 105 | 1.65 × 106 | 6.67 × 102 | 2.8 ± 0.36 | 18 |
| Enterococcus spp. | 1.08 × 105 | 5.05 × 105 | 1.50 × 103 | 2.7 ± 0.21 | 18 | |
| F-specific coliphages | 4.64 × 104 | 2.53 × 105 | 9.00 × 103 | 2.5 ± 0.15 | 9 | |
| Somatic coliphages | 7.28 × 103 | 1.15 × 104 | 1.00 × 103 | 3.2 ± 0.05 | 9 | |
The mean concentrations of the FIB and coliphages in the effluent of PSBR-L were slightly lower than those in the effluent of PSBR-H and exhibited a statistically significant difference, with p-values of 8.72 × 10−3, 5.07 × 10−3, 4.49 × 10−4, and 6.11 × 10−6 for E. coli, Enterococcus spp., F-specific and somatic coliphages, respectively. The PSBR-L had a higher concentration of chlorophyll and more diverse algae composition,17 which may contribute to the removal of FIB and viruses. E. coli and Enterococcus spp. removal may be impacted by the secondary metabolites generated by algae, including fatty acids, peptides, polysaccharides, and alkaloids.29 Based on 18S amplicon sequencing analysis,17 two species found in the PSBR, Scenedesmus spp. and Chlorella spp., are well-known to have robust antibacterial activity against both Gram-negative and Gram-positive bacteria.29 The relative abundance of Chlorella spp. ranged from 8–12% for PSBR-L and 1–6% for PSBR-H,17 while the relative abundance of Desmodesmus spp. was 32% and 74% for PSBR-L and PSBR-H, respectively.17 The air flow rate significantly affects the relative abundance of the dominant microalgae species (Chlorella spp. and Desmodesmus spp.). Increased aeration enhances oxygen availability and influences microbe–algae interactions, which are essential for nutrient cycling and pathogen removal. The bactericidal activity of Chlorella spp. and Desmodesmus spp. can be attributed to their production of reactive oxygen species and organic compounds, which contribute to the overall pathogen removal efficiency of the photobioreactor system.
Algae in aquatic environments have been reported to trap fine particles, which might include pathogenic bacteria (Fig. S4†). Filamentous algae might produce exopolysaccharides, which facilitate the adhesion of microbes to the algal filaments. This process is considered to be one of the potentially significant mechanisms for pathogen removal from wastewater in PSBR. Additionally, filamentous algae have been observed to harbor enteric bacteria, suggesting that these bacteria can survive for extended periods, sustained by algal exudates and shielded from UV radiation.30
The log10 removals of F-specific and somatic coliphages in PSBR-L were higher than that those in PSBR-H. There was a significant difference between the log10 removal of E. coli, F-specific and somatic coliphages between the two PSBRs, with p-values of 8.65 × 10−3, 1.17 × 10−3, 8.80 × 10−5, respectively. However, there was no significant difference in Enterococcus spp. removal between the two PSBRs (p-value = 5.98 × 10−2).
One of the potential removal mechanisms of indicators in the PSBRs is biological predation by protozoa organisms. 18S rRNA sequencing analysis illustrated an increase in the abundance of parasites such as rotifers and ciliated taxa during the operation of these algal–bacterial granular PSBRs.17 Predation might affect coliphages and FIB, and a further biological removal mechanism might occur for FIB, namely, cell lysis due to the use of E. coli or Enterococcus spp. as potential hosts for bacteriophage replication.31
Previous studies have demonstrated the efficacy of algae-based and other wastewater treatment systems for pathogenic indicator removal (Table 2). For example, Espinosa et al.,32 used high-rate algal ponds (HRAPs) for the post-treatment of an up-flow anaerobic sludge blanket reactor (UASB) effluent and achieved 4.06![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 removal of E. coli, which is higher than that in the current study. However, the present study achieved higher log10 removals of FIB and coliphages compared to other HRAP studies.32–35 The higher FIB and coliphage log10 removal units in the present study could be attributed to the better-controlled environmental conditions, which allowed for optimal growth and activity of the algal–bacterial consortia.36 Furthermore, the granular structure in the PSBRs facilitated the formation of biofilms, which effectively absorbed and removed pathogens from the wastewater. The high surface-area-to-volume ratio of the granules provided ample opportunities for adsorption and subsequent inactivation or removal of FIB and coliphages. Additionally, the variations in influent strength, with the possibility of a lower strength in the present study compared to other field studies, may have influenced the observed results.
log10 removal of E. coli, which is higher than that in the current study. However, the present study achieved higher log10 removals of FIB and coliphages compared to other HRAP studies.32–35 The higher FIB and coliphage log10 removal units in the present study could be attributed to the better-controlled environmental conditions, which allowed for optimal growth and activity of the algal–bacterial consortia.36 Furthermore, the granular structure in the PSBRs facilitated the formation of biofilms, which effectively absorbed and removed pathogens from the wastewater. The high surface-area-to-volume ratio of the granules provided ample opportunities for adsorption and subsequent inactivation or removal of FIB and coliphages. Additionally, the variations in influent strength, with the possibility of a lower strength in the present study compared to other field studies, may have influenced the observed results.
| Pathogen Indicator | Treatment system | Size/scale | Source of wastewater | Influent conc. log10 pathogen 100 mL−1* | log10 removal | Ref. |
|---|---|---|---|---|---|---|
| E. coli | UASB | Pilot-scale (343 L) | Domestic sewage | 7.84 ± 1.1 | 1.09 | 32 |
| HRAP | Two HRAPs, each 205 L | Effluent from UASB | 4.69 ± 0.63 | 4.06 | 32 | |
| HRAP | Two HRAPs, each 860 L | Domestic wastewater | 6.7 ± 0.42 | 1.77 | 33 | |
| HRAP | Loop raceway 30 m length and 2.5 m width | Septic tank treated effluent | 6.35 ± 0.37 | 1.75 | 34 | |
| HRAP | Loop raceway 30 m length and 2.5 m width | Facultative pond treated effluent | 3.71 ± 0.53 | 2.75 | 34 | |
| Horizontal flow constructed wetlands (HFCW) | 470–1800 m2 | Decentralized wastewater | 7.45 ± 0.21 | 2.70 | 37 | |
| Vertical flow constructed wetlands (VFCW) | 16–90 m2 | Decentralized wastewater | 5.8 ± 0.7 | 3.35 | 37 | |
| Biological sand filters (BSF) | 26 m2 | Decentralized wastewater | 5.7 ± 0.28 | 4.12 | 37 | |
| Biofilters (BF) | 50 m2 | Decentralized wastewater | 5.75 ± 0.49 | 4.06 | 37 | |
| CAS | Full-scale | Pre-treated effluent (domestic wastewater) | 6.85 ± 0.44 | 2.3 | 24 | |
| Membrane bioreactor (MBR) | Full-scale | Pre-treated effluent (domestic wastewater) | 6.85 ± 0.44 | 6.8 | 24 | |
| CAS | 27 m3 | Domestic wastewater | — | 3.2 | 38 | |
| PSBR-L | Lab-scale 2.5 L | Domestic wastewater | 7.41 ± 0.33 | 3.2 | Current study | |
| PSBR-H | Lab-scale 2.5 L | Domestic wastewater | 7.41 ± 0.33 | 2.8 | Current study | |
| Enterococcus spp. | HFCW | 470–1800 m2 | Decentralized wastewater | 6.0 ± 0.28 | 2.3 | 37 |
| VFCW | 16–90 m2 | Decentralized wastewater | 5.23 ± 1.44 | 2.41 | 37 | |
| BSF | 26 m2 | Decentralized wastewater | 4.9 ± 0.98 | 2.91 | 37 | |
| BF | 50 m2 | Decentralized wastewater | 5.4 ± 0.28 | 3.16 | 37 | |
| CAS | Full-scale | Pre-treated effluent (domestic wastewater) | 5.90 ± 0.22 | 1.4 | 24 | |
| MBR | Full-scale | Pre-treated effluent (domestic wastewater) | 5.90 ± 0.22 | 5.8 | 24 | |
| CAS | 27 m3 | Domestic wastewater | — | 3.2 | 38 | |
| PSBR-L | Lab-scale 2.5 L | Domestic wastewater | 7.03 ± 0.23 | 2.9 | Current study | |
| PSBR-H | Lab-scale 2.5 L | Domestic wastewater | 7.03 ± 0.23 | 2.7 | Current study | |
| F-specific coliphages | UASB | Pilot-scale (343 L) | Raw sewage | 5.21 ± 0.39 | 0.56 | 32 |
| HRAP | Two HRAPs, each 205 L | Effluent from UASB | 4.87 ± 0.59 | 1.70 | 32 | |
| HRAP1 | Loop raceway 30 m length and 2.5 m width | Septic tank treated effluent | 3.88 ± 0.5 | 0.07 | 35 | |
| HRAP2 | Loop raceway 30 m length and 2.5 m width | Effluent from HRAP2 | 2.43 ± 1.06 | 0.24 | 35 | |
| CAS | Full-scale | Pre-treated effluent (domestic wastewater) | 5.94 ± 0.43 | 3.1 | 24 | |
| MBR | Full-scale | Pre-treated effluent (domestic wastewater) | 5.94 ± 0.43 | 1.7 | 24 | |
| CAS | 27 m3 | Domestic wastewater | — | 3.5 | 38 | |
| PSBR-L | Lab-scale 2.5 L | Domestic wastewater | 6.74 ± 0.27 | 2.9 | Current study | |
| PSBR-H | Lab-scale 2.5 L | Domestic wastewater | 6.74 ± 0.27 | 2.5 | Current study | |
| Somatic coliphages | UASB | Pilot-scale (343 L) | Raw sewage | 5.67 ± 0.53 | 0.40 | 32 |
| HRAP | Two HRAPs, each 205 L | Effluent from UASB | 5.28 ± 0.53 | 1.15 | 32 | |
| CAS | Full-scale | Pre-treated effluent (domestic wastewater) | 6.02 ± 0.41 | 2.7 | 24 | |
| MBR | Full-scale | Pre-treated effluent (domestic wastewater) | 6.02 ± 0.41 | 2.7 | 24 | |
| PSBR-L | Lab-scale 2.5 L | Domestic wastewater | 6.16 ± 0.36 | 3.2 | Current study | |
| PSBR-H | Lab-scale 2.5 L | Domestic wastewater | 6.16 ± 0.36 | 3.1 | Current study |
We also compared the FIB and coliphage removal of the PSBRs with those of other biological wastewater treatment systems (Table 2). The PSBRs in the current study demonstrated competitive performance in pathogen removal, particularly for coliphages. For F-specific coliphages, PSBR-L achieved a 2.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 removal, while PSBR-H achieved a 2.5
log10 removal, while PSBR-H achieved a 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 removal, both of which exceeded the membrane bioreactor (MBR) system rate of 1.7
log10 removal, both of which exceeded the membrane bioreactor (MBR) system rate of 1.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10.24 For somatic coliphages, PSBR-L achieved a 3.2
log10.24 For somatic coliphages, PSBR-L achieved a 3.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 removal, and PSBR-H achieved a 3.1
log10 removal, and PSBR-H achieved a 3.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 removal, which were comparable to those of the conventional activated sludge (CAS) and MBR systems, which demonstrated a 2.7
log10 removal, which were comparable to those of the conventional activated sludge (CAS) and MBR systems, which demonstrated a 2.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 removal.24 The MBRs achieved even higher removal efficiencies for E. coli and Enterococcus spp.24,38 The performance of the MBRs can be attributed to the physical separation provided by the membrane filtration system, which effectively removes pathogens and other contaminants through size exclusion and adsorption onto the membrane surface. Moreover, MBRs operate with an extended solid retention time (SRT), allowing for the development of a more diverse and abundant microbial community capable of effectively degrading organic matter and inactivating pathogens. Although PSBRs may not achieve the same level of bacterial removal as MBRs, they perform competitively and frequently more effectively than many other systems, particularly in coliphage removal. This suggests that PSBRs could be advantageous in situations in which the costs or operational complexities of MBRs are prohibitive, particularly when viral pathogen removal is a major concern.
log10 removal.24 The MBRs achieved even higher removal efficiencies for E. coli and Enterococcus spp.24,38 The performance of the MBRs can be attributed to the physical separation provided by the membrane filtration system, which effectively removes pathogens and other contaminants through size exclusion and adsorption onto the membrane surface. Moreover, MBRs operate with an extended solid retention time (SRT), allowing for the development of a more diverse and abundant microbial community capable of effectively degrading organic matter and inactivating pathogens. Although PSBRs may not achieve the same level of bacterial removal as MBRs, they perform competitively and frequently more effectively than many other systems, particularly in coliphage removal. This suggests that PSBRs could be advantageous in situations in which the costs or operational complexities of MBRs are prohibitive, particularly when viral pathogen removal is a major concern.
3.2. Particle association of fecal indicator bacteria and coliphages
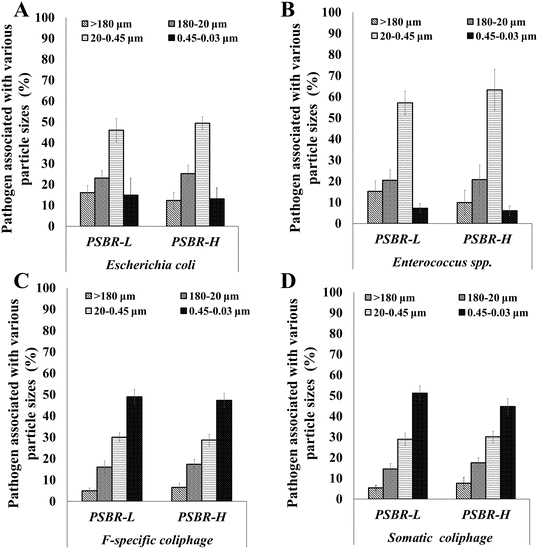
E. coli and Enterococcus spp. demonstrated the highest association with the particles in the size fraction 20–0.45 μm (46% and 57% for PSBR-L and 49% and 63% for PSBR-H), followed by the 180–20 μm, >180 μm, and 0.45–0.03 μm size fractions (Fig. 1). Prior study showed that 95% of FIB were attached to particles in the 30–0.45 μm size range, while less than 5% were retained on filters with particles >30 μm.39 Similarly, a study by Walters et al.40 illustrated that 91% of E. coli and 83% of Enterococcus spp. were associated with particle sizes ≤12 μm in the water column. There were no significant differences in the concentrations of E. coli and Enterococcus spp. among the 180–20 μm, >180 μm, and 0.45–0.03 μm particle size fractions for either reactor (p-value > 0.05).F-specific and somatic coliphages exhibited the highest concentrations on the 0.45–0.03 μm size fraction (48% and 51% for PSBR-L, and 47% and 44% for PSBR-H) compared to the particles retained on the other three filters. For both reactors, F-specific and somatic coliphages demonstrated a significant difference between the 20–0.45 μm and 180–20 μm size fractions. The current study highlights that most FIB were retained on filters with a 0.45 μm pore size, while coliphages were mainly retained on filters with pore sizes as small as 0.03 μm. Coliphages are smaller than bacteria and can pass through the pores of a 0.45 μm membrane unless they adsorb to the membrane or are associated with larger particles. Additionally, FIB and coliphages were not detected in the filtrate of the 0.03 μm membrane (<0.03 μm).
The association of bacterial cells with particles protects the cells from environmental stressors and provides them with organic matter and/or nutrients, enabling their survival and growth.41 As Chahal et al.16 demonstrated, larger particles settle due to gravity. The larger size of E. coli and Enterococcus spp. (0.5–5 μm) explains their retention on the 0.45 μm membrane.
On the other hand, coliphages, whose sizes range between 24–200 nm, are considerably smaller than bacteria and are typically associated with much smaller particles.16,42 The association of coliphages with particles is influenced by environmental factors, such as the pH, which affects the isoelectric point of their capsids. The significant portion (44–51%) of the viruses retained on membrane filters with a pore size of 0.03 μm indicated that the virus could remain in the liquid effluent after treatment, suggesting that their functional hydrodynamic diameter would be larger than that of the free coliphage particles.25 The functional hydrodynamic diameter phenomenon, known as apparent size, refers to the size of particles in fluid dispersions, which can differ from their individual size when they are associated with other materials and aggregates.43 Bacteria may be drawn toward nutrients released from the surfaces of various particles, leading them to migrate, attach, and ultimately colonize the surfaces of those particles.16
The absence of a difference in the 0.45–0.03 μm fraction between the two PSBRs, as shown in Fig. 1C, can be attributed to the similar operating conditions. Both reactors were exposed to comparable environmental parameters, such as temperature, light intensity, and nutrient availability. Although the differing aeration flow rates of PSBR-L and PSBR-H induced different dynamics in their microbial communities, no significant differences in the size or the settling ability of the granules of the two reactors were observed. The average particle sizes of PSBR-L and PSBR-H were 0.29 (SD = 0.07) mm and 0.25 (SD = 0.1) mm, respectively.17 Although the average particle size of PSBR-L was higher than that of PSBR-H, no significant differences were observed over long-term operation (P > 0.05). The size distribution indicated that over 80% of the particles in PSBR-L and PSBR-H were within the 0.1–0.5 mm range during long-term operation. These observations highlight the complex interactions between the operational parameters and granule morphology in PSBRs. 45.6% and 35.9% of the granules had a diameter of over 0.2 mm in PSBR-L and PSBR-H, respectively, while these granules contributed less than 20% of the FIB and coliphage indicator removal in both reactors. This signals that the flocs with smaller sizes (<0.2 mm) may have exhibited higher microbial adsorption capacity, which can be attributed to their higher specific surface area (m2 m−3). This increased surface area enhances the mass transfer rate of substrates and promotes more effective microbial adsorption capacity, contributing significantly to the removal of FIB and coliphages.
3.3. Superoxide production in PSBRs
The superoxide anion production was monitored from Day 130 to the end of the experiment. The average concentrations of superoxide in PSBR-L and PSBR-H were 0.179 (SD = 0.024) mmol L−1 and 0.206 (SD = 0.011) mmol L−1, respectively, which were nearly six times higher than those in the influent. The elevated aeration in PSBR-H resulted its having significantly higher ROS concentrations than PSBR-L. The increased airflow rate in PSBR-H enhanced the dissolved oxygen levels, stimulating metabolic activity and leading to increased production of ROS, such as superoxide and hydrogen peroxide.44,45 The superoxide concentrations varied with the operation time, from 0.110 to 0.213 mmol L−1 and 0.194 to 0.225 mmol L−1 for PSBR-L and PSBR-H, respectively. Ugya et al.46 reported superoxide production values of 0.261 (SD = 0.039) and 0.251 (SD = 0.148) mmol L−1 in Chlorella vulgaris and Aphanizomenon flos-aquae dominated biofilms, which are comparable to the high superoxide accumulation observed for PSBR-L and PSBR-H in the current study. The significant variation observed during treatment could be attributed to the complex microbial community, which stimulated superoxide generation through synergistic relationships between the algal–bacterial granules and endogenous microorganisms in real wastewater.47The FIB and coliphage concentrations showed relatively weak positive relationships with the ROS production in the reactors (Table S2†). Although no statistically significant correlations were found, these investigations contribute to our understanding of the complex interactions within the treatment process. The complex microbial ecologies lead to intricate interactions influencing reactive oxygen species production and pathogen indicators. While ROS can damage lipids, proteins, and DNA, leading to cell death, there is no direct evidence yet of their selectivity towards certain microorganisms over others. Previous studies revealed that algal–bacterial granules serve as inducible defenses against pathogens in wastewater treatment systems via ROS.48
Recent research shows that microalgae in wastewater produce antibacterial ROS under stresses such as nutrient deprivation.49 Moreover, ROS could suppress the growth of pathogens retained in the granules through oxidative damage. The treatment process may involve mechanisms that consume ROS or result in ROS-independent inactivation of pathogens.50 Further investigations are needed to understand the specific mechanisms governing ROS dynamics and their impact on pathogen inactivation.
3.4. Impact of light conditions and exposure to wastewater on ROS production in algal–bacterial consortia
Time-phased experiments were set up in real wastewater and synthetic wastewater for 48 h to examine the kinetics of ROS production. The initial algal–bacterial biomass concentration was standardized to ensure consistency between experiments, with an inoculum concentration of 1.2 × 107 cells per mL, while the mixed liquor suspended solids concentrations were 780 ± 53 mg L−1. The superoxide concentrations produced in real wastewater at 48 h were 0.159 (SD = 0.012) mmol L−1 and 0.124 (SD = 0.014) mmol L−1 under light and dark conditions, respectively. The ROS concentrations in synthetic wastewater were 0.067 (SD = 0.007) mmol L−1 and 0.026 (SD = 0.003) mmol L−1 under light and dark conditions, respectively (Fig. 2). | ||
| Fig. 2 Superoxide production profiles in the photobioreactors and raw wastewater during the stage 2 experimental period of reactor operation. | ||
Both the first-order model and the Gompertz model fit the experimental data for ROS production in synthetic wastewater well (Fig. 3), with R2 values of 0.98. However, in real wastewater, the Gompertz model demonstrated a better fit to the experimental data than the first-order reaction model (Fig. 3 and Table 3), with R2 values of 0.97 and 0.90, respectively. The reaction rate coefficients for real wastewater obtained from both models were higher than those for synthetic wastewater (Table 3), indicating that the complex substrates in real wastewater promoted superoxide production.
| Media | First-order reaction model | Gompertz model | ||||||
|---|---|---|---|---|---|---|---|---|
| a (mmol L−1) | b (mmol L−1) | k 1 (h−1) | R 2 | A (mmol L−1) | B (mmol L−1) | k 2 (h−1) | R 2 | |
| Real wastewater | 0.67 | 0.55 | 0.002 | 0.90 | 0.16 | −0.96 | 0.06 | 0.97 |
| Synthetic wastewater | 0.67 | 0.65 | 0.002 | 0.98 | 0.09 | 0.43 | 0.04 | 0.98 |
Significant amounts of ROS were produced under light conditions. Light-induced ROS production in algal–bacterial consortia is mainly due to algae photosynthesis. In a photobioreactor, light-induced ROS from algae occurs via generation of electrons during photosynthesis, forming superoxide radicals in the thylakoid membrane.51 Simultaneously, oxygen is produced as a byproduct when water molecules split. Several studies have shown that exposure to light, especially high-intensity visible light and ultraviolet radiation, can trigger oxidative stress and ROS accumulation in photosynthetic organisms, including microalgae and cyanobacteria.52
Exposure to real wastewater induced significantly higher ROS production under both light and dark conditions (Fig. 2 and 3). Algae and bacteria have mechanisms, including enzymes like superoxide dismutase and catalase, to detoxify ROS and protect cells.53,54 These insights highlight how the wastewater composition profoundly impacts algal–bacterial oxidative stress and ROS responses.55,56
3.5. Microbial community analysis
The influent samples had a diverse microbial population, with 38 distinct genera that were not detected in the other stages. In PSBR-L, there was a change in community composition, with 6 distinct genera detected. At a low airflow rate, the complex multilayered structure of the algal–bacterial granules provides varied microenvironments for different bacterial species, enabling the maintenance of microbial diversity and potentially improving the system's capacity to eliminate a wide range of particle-associated fecal indicators.
Interestingly, the PSBR-L effluent shared 50 genera with the sludge in the PSBR-L reactor, indicating that a significant portion of the microbial community was carried over from the PSBR-L reactor to the effluent. Similarly, the effluent-PSBR-H stage shared 17 genera with the PSBR-H stage, but also had 5 unique genera. The presence of shared genera across multiple stages indicates that certain microbial populations were able to adapt and thrive under the varying conditions encountered during the treatment process.
Alpha diversity and richness were evaluated using the Shannon and Chao1 indices (Table S1†). The analysis indicated that all samples had diverse and complex microbial communities, as supported by high OTU counts (32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 847–50
847–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 241). The Shannon diversity indices ranged from 4.97 to 5.29. The influent sample had the highest OTU count of 50
241). The Shannon diversity indices ranged from 4.97 to 5.29. The influent sample had the highest OTU count of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 241 and richness. The sludge in PSBR-L showed the highest Shannon diversity, while the PSBR-H sludge sample had the lowest estimate.
241 and richness. The sludge in PSBR-L showed the highest Shannon diversity, while the PSBR-H sludge sample had the lowest estimate.
Flavobacterium spp. and Solibacillus spp., which have been identified as significant players in potential pathogen removal, showed relative abundances of less than 1% in all samples.65,66Flavobacterium causes fish diseases, but also exhibits antagonistic properties against pathogens. Flavobacterium can inhibit the growth of human pathogen bacteria by producing antibiotics, bacteriocins, or siderophores.67 Earlier studies revealed that Flavobacterium, an EPS-secreting bacterium with strong self-agglomeration ability, could assist in the removal of nitrogen and phosphate as a denitrifying phosphate-accumulating organism.68,69 On the other hand, Solibacillus spp. exhibits antimicrobial activity, indicating its potential contribution to the elimination of particle-associated fecal indicators.66 Noteworthily, the presence of genera such as Escherichia, Enterococcus spp., Streptococcus spp., Aeromonas spp., and Clostridium spp., was detected in the influent, PSBRs, and their effluents with relative abundances below 1.0%.
Algal–bacterial granules make contributions to wastewater treatment, including the elimination of particle-associated fecal indicators. These granules facilitate symbiotic relationships within the microbial community, enhancing the capture and inactivation of fecal indicators.70 For instance, the interaction between cyanobacteria (as Nodosilinea spp. and Leptolyngbya spp.) and the coexistence of nitrifying and denitrifying bacteria within the granules may have permitted efficient nitrogen cycling and elimination of FIB.71,72
Furthermore, based on 18S amplicon sequencing analysis,17 the abundance of algae as Chlorella spp., Desmodesmus spp. and Scenedesmus spp. are vital for the effective decomposition of complex organic matter as well as pathogen removal. The effective exchange of O2 and CO2 between algae and bacteria in these interactions is vital for enhancing the removal efficiency of organics in the photobioreactor. The impact of the symbiotic interactions and the metabolic activity of the algal–bacterial granules on the environmental conditions of the photobioreactor, such as pH, might impact the elimination or inactivation rates of the FIB and coliphages. These microbial interactions and species-specific contributions are closely linked to the observed trends in particle association, pathogen removal, and ROS production, emphasizing the critical role of community structure in determining photobioreactor performance. However, further investigation is required to fully understand the mechanisms of pathogen removal using algal–bacterial granules and to characterize the nutrient dynamics.
4. Conclusion
PSBRs with different airflow rates effectively removed fecal bacteria and viral indicators from wastewater, and it was revealed that the airflow rate had no significant impact on the removal of pathogen indicators. FIB was primarily retained on 0.45 μm pore size membranes, while viruses were retained on 0.03 μm pore size membranes. Association with particles plays a crucial role in the persistence and transportation of FIB in the environment. The superoxide levels produced in the PSBRs were higher than in the influent due to complex microbial interactions during treatment. Furthermore, under light conditions, the levels of superoxide in real wastewater were significantly higher than those in synthetic wastewater. The microbial community structure shifted at different stages of the treatment in the photobioreactors, while the key nutrient-removal genera remained consistent. PSBRs have the potential to produce high-quality effluents with reduced pathogens, indicating promising applications in water reuse. Further research is necessary to optimize the community structure–function relationships in photobioreactors.Data availability
Sequencing data has been deposited to the NCBI Sequence Read Archive under BioProject accession number: PRJNA1100726.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based upon work supported by the Penn State Institute of Energy and the Environment (IEE) seed grant program. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the sponsors.References
- J. Ding, Y. Meng, S. Lu, Y. Peng, W. Yan, W. Li, J. Hu, T. Ye, Y. Zhong and H. Zhang, The Treatment of Aquaculture Wastewater with Biological Aerated Filters: From the Treatment Process to the Microbial Mechanism, Toxics, 2023, 11, 478 CrossRef CAS PubMed.
- S. Li, S. Xie, Y. Yang, X. Yang, Y. Lu, L. Luo, S. Chen and T. Luan, Inhibition mechanisms of high salinities on flocculation of marine Algal-Mycelial pellets, Curr. Res. Biotechnol., 2024, 7, 100222 CrossRef CAS.
- M. Z. Khan, P. K. Mondal and S. Sabir, Aerobic granulation for wastewater bioremediation: a review, Can. J. Chem. Eng., 2013, 91, 1045–1058 CrossRef CAS.
- M.-K. H. Winkler, C. Meunier, O. Henriet, J. Mahillon, M. E. Suárez-Ojeda, G. Del Moro, M. De Sanctis, C. Di Iaconi and D. G. Weissbrodt, An integrative review of granular sludge for the biological removal of nutrients and recalcitrant organic matter from wastewater, Chem. Eng. J., 2018, 336, 489–502 CrossRef CAS.
- É. Sylvestre, S. Dorner, J.-B. Burnet, P. Smeets, G. Medema, P. Cantin, M. Villion, C. Robert, D. Ellis and P. Servais, Changes in Escherichia coli to enteric protozoa ratios in rivers: Implications for risk-based assessment of drinking water treatment requirements, Water Res., 2021, 205, 117707 CrossRef PubMed.
- C. A. Baker, G. Almeida, J. A. Lee and K. E. Gibson, Pathogen and surrogate survival in relation to fecal indicator bacteria in freshwater mesocosms, Appl. Environ. Microbiol., 2021, 87, e00558-00521 CrossRef PubMed.
- P. Teixeira, D. Salvador, J. Brandão, W. Ahmed, M. J. Sadowsky and E. Valério, Environmental and adaptive changes necessitate a paradigm shift for indicators of fecal contamination, Microbiol. Spectrum, 2020, 8 DOI:10.1128/microbiolspec.erv-0001-2019.
- M. A. El-Liethy and A. L. K. Abia, Some bacterial pathogens of public health concern in water and wastewater: An African perspective, Current Microbiological Research in Africa: Selected Applications for Sustainable Environmental Management, 2020, pp. 1–27 Search PubMed.
- E. Li, F. Saleem, T. A. Edge and H. E. Schellhorn, Biological indicators for fecal pollution detection and source tracking: A review, Processes, 2021, 9, 2058 CrossRef CAS.
- A. Bhatt, P. Arora and S. K. Prajapati, Occurrence, fates and potential treatment approaches for removal of viruses from wastewater: a review with emphasis on SARS-CoV-2, J. Environ. Chem. Eng., 2020, 8, 104429 CrossRef CAS PubMed.
- P. Dange, S. Gawas, S. Pandit, L. Mekuto, P. K. Gupta, P. Shanmugam, R. Patil and S. Banerjee, in An Integration of Phycoremediation Processes in Wastewater Treatment, Elsevier, 2022, pp. 135–154 Search PubMed.
- F. Thevenon and F. Thevenon, Sustainable wastewater and sanitation systems: Health, environment and governance challenges: The case for human rights-based policy reform in alternative waste-water management strategies, WaterLex and the UN Environment Global Wastewater Initiative, Geneva, Switzerland, 2017 Search PubMed.
- K. K. Kesari, R. Soni, Q. M. S. Jamal, P. Tripathi, J. A. Lal, N. K. Jha, M. H. Siddiqui, P. Kumar, V. Tripathi and J. Ruokolainen, Wastewater treatment and reuse: a review of its applications and health implications, Water, Air, Soil Pollut., 2021, 232, 1–28 CrossRef.
- J. P. Sheets, L. Yang, X. Ge, Z. Wang and Y. Li, Beyond land application: Emerging technologies for the treatment and reuse of anaerobically digested agricultural and food waste, Waste Manage., 2015, 44, 94–115 CrossRef CAS PubMed.
- K. Yang, L. Li, Y. Wang, S. Xue, Y. Han and J. Liu, Airborne bacteria in a wastewater treatment plant: emission characterization, source analysis and health risk assessment, Water Res., 2019, 149, 596–606 CrossRef CAS PubMed.
- C. Chahal, B. Van Den Akker, F. Young, C. Franco, J. Blackbeard and P. Monis, Pathogen and particle associations in wastewater: significance and implications for treatment and disinfection processes, Adv. Appl. Microbiol., 2016, 97, 63–119 CAS.
- T. Zhang, W. M. M. El-Sayed, J. Zhang, L. He, M. A. Bruns and M. Wang, Insight into the Impact of Air Flow Rate on Algal-Bacterial Granules: Reactor Performance, Hydrodynamics by Computational Fluid Dynamics (CFD) and Microbial Community Analysis, Chem. Eng. J., 2024, 499, 156275 CrossRef CAS.
- E. W. Rice, R. B. Baird and A. D. Eaton, Standard methods for the examination of water and wastewater, 2012 Search PubMed.
- M. E. Verbyla, M. Fani and B. Walker, Pathogen Removal Credits for Wastewater Treatment: Guidance for Study Plans and Reporting, Final Report, 2023 Search PubMed.
- J. Greaves, D. North and K. Bibby, Particle association and size fraction of molecular viral fecal pollution indicators in wastewater, Environ. Sci.: Water Res. Technol., 2022, 8, 1814–1821 RSC.
- E. Symonds, M. Verbyla, J. Lukasik, R. Kafle, M. Breitbart and J. Mihelcic, A case study of enteric virus removal and insights into the associated risk of water reuse for two wastewater treatment pond systems in Bolivia, Water Res., 2014, 65, 257–270 CrossRef CAS PubMed.
- M. Hayyan, M. A. Hashim and I. M. AlNashef, Superoxide ion: generation and chemical implications, Chem. Rev., 2016, 116, 3029–3085 CrossRef CAS PubMed.
- C. Liu, Y. Cui, X. Li and M. Yao, microeco: an R package for data mining in microbial community ecology, FEMS Microbiol. Ecol., 2021, 97, fiaa255 CrossRef CAS PubMed.
- G. De Luca, R. Sacchetti, E. Leoni and F. Zanetti, Removal of indicator bacteriophages from municipal wastewater by a full-scale membrane bioreactor and a conventional activated sludge process: implications to water reuse, Bioresour. Technol., 2013, 129, 526–531 CrossRef CAS PubMed.
- B. R. McMinn, N. J. Ashbolt and A. Korajkic, Bacteriophages as indicators of faecal pollution and enteric virus removal, Lett. Appl. Microbiol., 2017, 65, 11–26 CrossRef CAS PubMed.
- K. Zhang and K. Farahbakhsh, Removal of native coliphages and coliform bacteria from municipal wastewater by various wastewater treatment processes: implications to water reuse, Water Res., 2007, 41, 2816–2824 CrossRef CAS PubMed.
- M. Henze, M. C. van Loosdrecht, G. A. Ekama and D. Brdjanovic, Biological wastewater treatment, IWA publishing, 2008 Search PubMed.
- B. Matthews, H. Stratton, S. Schreoder and S. Toze, Pathogen detection methodologies for wastewater and reservoirs, Urban water security research alliance technical report, 2010, vol. 32 Search PubMed.
- C. Falaise, C. François, M.-A. Travers, B. Morga, J. Haure, R. Tremblay, F. Turcotte, P. Pasetto, R. Gastineau and Y. Hardivillier, Antimicrobial compounds from eukaryotic microalgae against human pathogens and diseases in aquaculture, Mar. Drugs, 2016, 14, 159 CrossRef PubMed.
- A. Beckinghausen, A. Martinez, D. Blersch and B. Z. Haznedaroglu, Association of nuisance filamentous algae Cladophora spp. with E. coli and Salmonella in public beach waters: impacts of UV protection on bacterial survival, Environ. Sci.: Processes Impacts, 2014, 16, 1267–1274 RSC.
- R. Young, Bacteriophage lysis: mechanism and regulation, Microbiol. Rev., 1992, 56, 430–481 CrossRef CAS PubMed.
- M. F. Espinosa, M. E. Verbyla, L. Vassalle, A. T. Rosa-Machado, F. Zhao, A. Gaunin and C. R. Mota, Reduction and partitioning of viral and bacterial indicators in a UASB reactor followed by high rate algal ponds treating domestic sewage, Sci. Total Environ., 2021, 760, 144309 CrossRef CAS PubMed.
- P. Chambonniere, J. Bronlund and B. Guieysse, Escherichia coli removal during domestic wastewater treatment in outdoor high rate algae ponds: long-term performance and mechanistic implications, Water Sci. Technol., 2020, 82, 1166–1175 CrossRef CAS PubMed.
- N. A. Buchanan, P. Young, N. J. Cromar and H. J. Fallowfield, Performance of a high rate algal pond treating septic tank effluent from a community wastewater management scheme in rural South Australia, Algal Res., 2018, 35, 325–332 CrossRef.
- H. J. Fallowfield, P. Young, M. J. Taylor, N. Buchanan, N. Cromar, A. Keegan and P. Monis, Independent validation and regulatory agency approval for high rate algal ponds to treat wastewater from rural communities, Environ. Sci.: Water Res. Technol., 2018, 4, 195–205 RSC.
- M. Verbyla, M. Von Sperling and Y. Maiga, Waste stabilization ponds, Global Water Pathogens Project. Part 4—Management of Risk from Excreta and Wastewater, 2017 Search PubMed.
- B. Adrados, C. A. Arias, L. M. Perez, F. Codony, E. Bécares, H. Brix and J. Morató, Comparison of removal efficiency of pathogenic microbes in four types of wastewater treatment systems in Denmark, Ecol. Eng., 2018, 124, 1–6 CrossRef.
- J. Ottoson, A. Hansen, B. Björlenius, H. Norder and T. Stenström, Removal of viruses, parasitic protozoa and microbial indicators in conventional and membrane processes in a wastewater pilot plant, Water Res., 2006, 40, 1449–1457 CrossRef CAS PubMed.
- H. C. Jeng, A. J. England and H. B. Bradford, Indicator organisms associated with stormwater suspended particles and estuarine sediment, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2005, 40, 779–791 CrossRef CAS PubMed.
- E. Walters, M. Graml, C. Behle, E. Müller and H. Horn, Influence of particle association and suspended solids on UV inactivation of fecal indicator bacteria in an urban river, Water, Air, Soil Pollut., 2014, 225, 1–9 CrossRef CAS.
- G. F. Alotaibi and M. A. Bukhari, Factors influencing bacterial biofilm formation and development, Am. J. Biomed. Res., 2021, 12, 617–626 CrossRef.
- K. Raveendran, M. Vaiyapuri, M. Benala, V. Sivam and M. R. Badireddy, Diverse infective and lytic machineries identified in genome analysis of tailed coliphages against broad spectrum multidrug-resistant Escherichia coli, Int. Microbiol., 2023, 26, 459–469 CrossRef CAS PubMed.
- B. Y. Shekunov, P. Chattopadhyay, H. H. Tong and A. H. Chow, Particle size analysis in pharmaceutics: principles, methods and applications, Pharm. Res., 2007, 24, 203–227 CrossRef CAS PubMed.
- A. Y. Ugya, H. Chen and Q. Wang, Microalgae biofilm system as an efficient tool for wastewater remediation and potential bioresources for pharmaceutical product production: an overview, Int. J. Phytorem., 2024, 26, 131–142 CrossRef CAS PubMed.
- C. A. Sepúlveda-Muñoz, I. de Godos and R. Muñoz, Wastewater Treatment Using Photosynthetic Microorganisms, Symmetry, 2023, 15, 525 CrossRef.
- Y. A. Ugya, D. u. B. Hasan, S. M. Tahir, T. S. Imam, H. A. Ari and X. Hua, Microalgae biofilm cultured in nutrient-rich water as a tool for the phycoremediation of petroleum-contaminated water, Int. J. Phytorem., 2021, 23, 1175–1183 CrossRef CAS PubMed.
- S. Ariyadasa, W. Taylor, L. Weaver, E. McGill, C. Billington and I. Pattis, Nonbacterial Microflora in Wastewater Treatment Plants: an Underappreciated Potential Source of Pathogens, Microbiol. Spectrum, 2023, 11, e00481-00423 Search PubMed.
- D. Nagarajan, D.-J. Lee, S. Varjani, S. S. Lam, S. I. Allakhverdiev and J.-S. Chang, Microalgae-based wastewater treatment–microalgae-bacteria consortia, multi-omics approaches and algal stress response, Sci. Total Environ., 2022, 845, 157110 CrossRef CAS PubMed.
- W. Xinjie, N. Xin, C. Qilu, X. Ligen, Z. Yuhua and Z. Qifa, Vetiver and Dictyosphaerium sp. co-culture for the removal of nutrients and ecological inactivation of pathogens in swine wastewater, J. Adv. Res., 2019, 20, 71–78 CrossRef CAS PubMed.
- A. B. Inuwa, Q. Mahmood, J. Iqbal, E. Widemann, S. Shafiq, M. Irshad, U. Irshad, A. Iqbal, F. Hafeez and R. Nazir, Removal of Antibiotic Resistance Genes, Class 1 Integrase Gene and Escherichia coli Indicator Gene in a Microalgae-Based Wastewater Treatment System, Antibiotics, 2022, 11, 1531 CrossRef CAS PubMed.
- X. Zheng, Z. Xu, J. Liu, Y. Luo, L. Gu, D. Zhao, S. Hu and X. Pan, Multiple roles of dissolved organic matter on typical engineered nanomaterials: environmental behaviors, pollutants removal and potential risks, Carbon Res., 2022, 1, 27 CrossRef CAS.
- Y. Astafyeva, M. Gurschke, M. Qi, L. Bergmann, D. Indenbirken, I. de Grahl, E. Katzowitsch, S. Reumann, D. Hanelt and M. Alawi, Microalgae and bacteria interaction—Evidence for division of diligence in the alga microbiota, Microbiol. Spectrum, 2022, 10, e00633-00622 Search PubMed.
- L. Jiang, Y. Li and H. Pei, Algal–bacterial consortia for bioproduct generation and wastewater treatment, Renewable Sustainable Energy Rev., 2021, 149, 111395 CrossRef CAS.
- S. F. Ahmed, M. Mofijur, T. A. Parisa, N. Islam, F. Kusumo, A. Inayat, I. A. Badruddin, T. Y. Khan and H. C. Ong, Progress and challenges of contaminate removal from wastewater using microalgae biomass, Chemosphere, 2022, 286, 131656 CrossRef CAS PubMed.
- U. S. Hyder, The Effect of pH and Hydraulic Retention Time on Production of Volatile Fatty Acids from Primary Sludge Anaerobic Fermentation, Doctoral dissertation, Toronto Metropolitan University, 2018 Search PubMed.
- C.-Y. Hsieh, Y.-C. Wu, S. Mudigonda, H.-U. Dahms and M.-C. Wu, Assessing the Effects of Ozonation on the Concentrations of Personal Care Products and Acute Toxicity in Sludges of Wastewater Treatment Plants, Toxics, 2023, 11, 75 CrossRef CAS PubMed.
- A. Rada-Ariza, C. Lopez-Vazquez, N. Van der Steen and P. Lens, in Algal Systems for Resource Recovery from Waste and Wastewater, IWA Publishing, 2023, pp. 31–52 Search PubMed.
- F. Cai, S. Li, H. Zhang, G. Yu and R. Li, Nodosilinea hunanesis sp. nov. (Prochlorotrichaceae, Synechococcales) from a freshwater pond in China based on a polyphasic approach, Diversity, 2022, 14, 364 CrossRef CAS.
- F. Chassagne, T. Samarakoon, G. Porras, J. T. Lyles, M. Dettweiler, L. Marquez, A. M. Salam, S. Shabih, D. R. Farrokhi and C. L. Quave, A systematic review of plants with antibacterial activities: A taxonomic and phylogenetic perspective, Front. Pharmacol., 2021, 11, 2069 Search PubMed.
- A. Lykov, A. Salmin, R. Gevorgiz, S. Zheleznova, L. Rachkovskaya, M. Surovtseva and O. Poveshchenko, Study of the Antimicrobial Potential of the Arthrospira platensis, Planktothrix agardhii, Leptolyngbya cf. ectocarpi, Roholtiella mixta nov., Tetraselmis viridis, and Nanofrustulum shiloi against Gram-Positive, Gram-Negative Bacteria, and Mycobacteria, Mar. Drugs, 2023, 21, 492 CrossRef CAS PubMed.
- S. A. Begmatov, A. Dorofeev, N. Pimenov, A. Mardanov and N. Ravin, High Efficiency of Removal of Pathogenic Microorganisms at Wastewater Treatment Plants in the City of Moscow, Microbiology, 2023, 92, 734–738 CAS.
- M. Yasir, Analysis of microbial communities and pathogen detection in domestic sewage using metagenomic sequencing, Diversity, 2020, 13, 6 CrossRef.
- S. Begmatov, A. G. Dorofeev, V. V. Kadnikov, A. V. Beletsky, N. V. Pimenov, N. V. Ravin and A. V. Mardanov, The structure of microbial communities of activated sludge of large-scale wastewater treatment plants in the city of Moscow, Sci. Rep., 2022, 12, 3458 CrossRef CAS PubMed.
- Q. Wang and J. He, Complete nitrogen removal via simultaneous nitrification and denitrification by a novel phosphate accumulating Thauera sp. strain SND5, Water Res., 2020, 185, 116300 CrossRef CAS PubMed.
- P. Maza-Márquez, M. Gallardo-Altamirano, F. Osorio, C. Pozo and B. Rodelas, Microbial indicators of efficient performance in an anaerobic/anoxic/aerobic integrated fixed-film activated sludge (A2O-IFAS) and a two-stage mesophilic anaerobic digestion process, Chemosphere, 2023, 139164 CrossRef PubMed.
- K. Parajuli, N. Fahim, S. Mumu, R. Palu and A. Mustafa, Antibacterial potential of Luidia clathrata (sea star) tissue extracts against selected pathogenic bacteria, PLoS One, 2023, 18, e0281889 CrossRef CAS PubMed.
- M. Ström-Bestor and T. Wiklund, Inhibitory activity of Pseudomonas sp. on Flavobacterium psychrophilum, in vitro, J. Fish Dis., 2011, 34, 255–264 CrossRef PubMed.
- Y. Li, Y. Xu, L. Liu, P. Li, Y. Yan, T. Chen, T. Zheng and H. Wang, Flocculation mechanism of Aspergillus niger on harvesting of Chlorella vulgaris biomass, Algal Res., 2017, 25, 402–412 CrossRef.
- A. Sengar, A. Aziz, I. H. Farooqi and F. Basheer, Development of denitrifying phosphate accumulating and anammox micro-organisms in anaerobic hybrid reactor for removal of nutrients from low strength domestic sewage, Bioresour. Technol., 2018, 267, 149–157 CrossRef CAS PubMed.
- C. Dang, Y. Zhang, M. Zheng, Q. Meng, J. Wang, Y. Zhong, Z. Wu, B. Liu and J. Fu, Effect of chlorine disinfectant influx on biological sewage treatment process under the COVID-19 pandemic: performance, mechanisms and implications, Water Res., 2023, 244, 120453 CrossRef CAS PubMed.
- A. Rosa-Masegosa, A. Rodriguez-Sanchez, S. Gorrasi, M. Fenice, A. Gonzalez-Martinez, J. Gonzalez-Lopez and B. Muñoz-Palazon, Microbial Ecology of Granular Biofilm Technologies for Wastewater Treatment: A Review, Microorganisms, 2024, 12, 433 CrossRef CAS PubMed.
- M. Pavlovska, I. Prekrasna, I. Parnikoza and E. Dykyi, Soil sample preservation strategy affects the microbial community structure, Microbes Environ., 2021, 36, ME20134 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ew00927d |
| This journal is © The Royal Society of Chemistry 2025 |