Revelation of adhesive proteins affecting cellular contractility through reference-free traction force microscopy†
Yingjun
Yang
 ae,
Kuankuan
Han
a,
Siyuan
Huang
ad,
Kai
Wang
a,
Yuchen
Wang
a,
Shukai
Ding
ae,
Kuankuan
Han
a,
Siyuan
Huang
ad,
Kai
Wang
a,
Yuchen
Wang
a,
Shukai
Ding
 a,
Le
Zhang
a,
Miao
Zhang
a,
Bingshe
Xu
aeg,
Shufang
Ma
a,
Le
Zhang
a,
Miao
Zhang
a,
Bingshe
Xu
aeg,
Shufang
Ma
 a,
Yongtao
Wang
a,
Yongtao
Wang
 *f,
Shengli
Wu
*c and
Xinlong
Wang
*f,
Shengli
Wu
*c and
Xinlong
Wang
 *b
*b
aMaterials Institute of Atomic and Molecular Science, School of Physics & Information Science, Shaanxi University of Science and Technology, Xi’an, P. R. China
bDepartment of Biomedical Engineering, Northwestern University, Evanston, IL, USA. E-mail: xinlong.wang@northwestern.edu
cHepatobiliary Surgery, The First Affiliated Hospital of Xi'an Jiaotong University, Xi’an, Shaanxi, China. E-mail: victorywu2000@163.com
dDepartment of Basic Medical Sciences, Sichuan Vocational College of Health and Rehabilitation, Zigong, China
eShanxi-Zheda Institute of Advanced Materials and Chemical Engineering, Taiyuan, P. R. China
fShanghai Engineering Research Center of Organ Repair, School of Medicine, Shanghai University, Shanghai, China. E-mail: yongtao_wang@shu.edu.cn
gLaboratory of Interface Science and Engineering in Advanced Materials, Taiyuan University of Technology, Taiyuan, China
First published on 28th February 2024
Abstract
Over the past few decades, the critical role played by cellular contractility associated mechanotransduction in the regulation of cell functions has been revealed. In this case, numerous biomaterials have been chemically or structurally designed to manipulate cell behaviors through the regulation of cellular contractility. In particular, adhesive proteins including fibronectin, poly-L-lysine and collagen type I have been widely applied in various biomaterials to improve cell adhesion. Therefore, clarifying the effects of adhesive proteins on cellular contractility has been valuable for the development of biomaterial design. In this study, reference-free traction force microscopy with a well-organized microdot array was designed and prepared to investigate the relationship between adhesive proteins, cellular contractility, and mechanotransduction. The results showed that fibronectin and collagen type I were able to promote the assembly of focal adhesions and further enhance cellular contraction and YAP activity. In contrast, although poly-L-lysine supported cell spreading and elongation, it was inefficient at inducing cell contractility and activating YAP. Additionally, compared with cellular morphogenesis, cellular contraction was essential for YAP activation.
Introduction
In vivo, cells are simultaneously affected by various soluble factors and biophysical stimuli from the extracellular matrix (ECM) or adjacent cells.1 Over the past few decades, the critical role played by these stimuli from the ECM in regulating cell behaviors through mechanotransduction has been revealed by numerous studies.2,3 Within mechanotransduction, cytoskeletons are remodeled and used for translating external stimuli into biochemical signals by sensing local changes in the ECM composition and mechanics.4 The remodeled cytoskeleton structures always result in changes in cellular contractility.5 Based on previous reports, cellular contractility is tightly related to various cellular functions and biological processes, including cellular alignments,6 chromatin condensation,7 proliferation,8 migration,9 determination of stem cell fate,10 embryogenesis,11 and epithelial scattering.12 Therefore, cellular contractility is considered the most important link between the external environment and intracellular biological processes. Thus, the chemical components and mechanical properties of biomaterials are designed and optimized to manipulate various cell behaviors through the regulation of cellular contractility associated mechanotransduction.13,14As one of the fundamental elements in mechanotransduction, connections between cells and adhesive proteins in the ECM are essential for the regulation of cell functions and tissue development.15 In particular, cells are initially anchored to adhesive proteins in the ECM through cadherin, integrin, or other transmembrane proteins.16,17 These cell adhesion anchors are able to transfer extracellular stimuli to their integrated cytoskeleton and induce cytoskeleton remodeling.18 In addition, adhesive proteins including fibronectin, poly-L-lysine, and collagen type I have been extensively applied to promote cell adhesion on material surfaces.19 Therefore, in order to provide essential information about the design of cell adhesive biomaterials, it is desirable to clarify the effects of adhesive proteins on cellular contractility. Unfortunately, even though external mechanical stimuli20,21 have been demonstrated to be efficient in manipulating cellular contractility, the effects of adhesive proteins on cell contraction remain unclear.
Additionally, since the determination of cellular contractility is critical for deciphering the biological processes of cells sensing local changes in the in situ microenvironment, the detection of cellular contractility has always been the goal of materials scientists and biologists. Initially, cellular contractility was visualized by observing the wrinkled deformation of the thin silicone membrane caused by cell contraction.22 Recently, techniques for the detection of cellular contractility have been explosively developed. For instance, traction force microscopy (TFM) with fluorescent particles and an elastic hydrogel has been broadly used in the detection of contractile force.23 In traditional TFM, fluorescent particles were randomly distributed on the elastic hydrogel. After cell culture, cellular contractility was detected through the calculation of particle displacement.24 However, traditional TFM still faces some limitations. Specifically, a reference image showing the original distribution of fluorescent particles was indispensable for the calculation of particle displacement.25 In this case, cells must be freshly cultured or removed for in situ observation. Because of this additional procedure, traditional TFM was difficult to detect the dynamic changes in cellular contractility. Subsequently, reference-free TFMs with well-organized patterns were developed. Typically, regularly arranged fluorescent dots or pillars formed the foundation for reference-free TFMs.26–29 These reference-free TFMs provided an approachable technique for sensing cellular contractility during cell growth. However, the tedious preparation process and the request of precious instruments including focused laser beam or electron-beam lithography limited their broad applications. Thus, it was necessary to develop a reference-free TFM with a simple preparation process and economical instruments.
In this research, a reference-free TFM was developed by using approachable photolithography and commercially available materials. The well-organized microdots of fluorescent photoresist were designed and prepared on an elastic polyacrylamide (PAA) gel. The displacement of the microdots was measured to calculate cell contractility. In addition, fibronectin, poly-L-lysine, and collagen type I were modified on the reference-free TFM to explore the effects of these adhesive proteins on cellular contractility.
Materials and methods
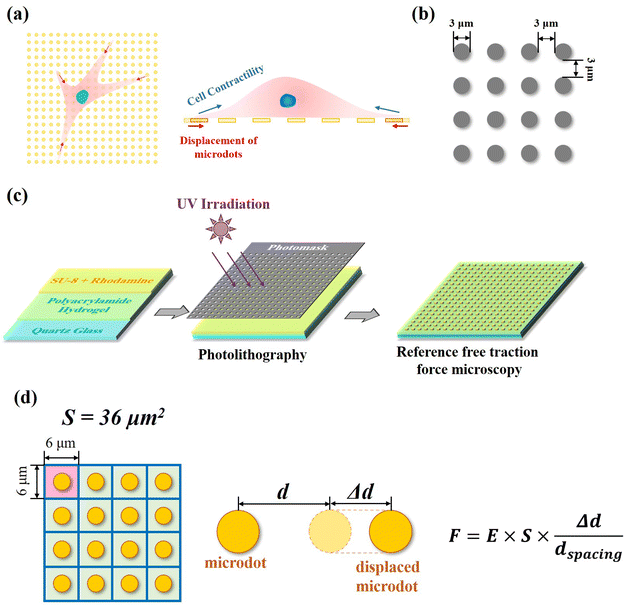
Preparation and characterization of reference-free traction force microscopy
In our reference-free TFM, the well-arranged microdot array was designed to detect cellular contractility through the measurement of microdot displacement (Fig. 1a). A photomask containing microdots with 3 μm in diameter and 3 μm spacing was designed and applied for photolithography (Fig. 1b). The reference-free TFM was prepared through photolithography using photoresist SU-8 and elastic polyacrylamide (PAA) gel. Specifically, acrylamide (Shanghai Aladdin Biochemical Technology Co., Ltd, Shanghai, China, 10% w/w) and N,N-methylene-bis-acrylamide (BIS, Shanghai Aladdin Biochemical Technology Co., Ltd, Shanghai, China, 0.03% w/w) were firstly dissolved in Milli-Q water and further degassed. Then, N,N,N,N-tetramethyl ethylenediamine (TEMED, Shanghai Aladdin Biochemical Technology Co., Ltd, Shanghai, China, 0.05% v/v) and ammonium persulfate (Shanghai Aladdin Biochemical Technology Co., Ltd, Shanghai, China, 0.05% v/v) were added to the above solution. Subsequently, as shown in Fig. 1c, 200 μL of the prepared solution was coated on a 10 mm × 10 mm quartz glass and further flattened by covering it with a cover slip. After polymerization for 30 min at room temperature, the samples were dried for another 24 h. After the preparation of elastic PAA gel on quartz slides, a mixture of photoresist (SU-8 2000.5, Microchem Laboratory, Newton, MA, USA) and rhodamine B (Shanghai Aladdin Biochemical Technology Co., Ltd, Shanghai, China, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 mixed in SU-8) was spin coated on the dried samples (1500 rpm, 30 s). After baking (95 °C, 3 min), the samples were exposed to UV irradiation through the designed photomask. Fluorescent SU-8 microdots on the dried PAA gel were obtained after the development process and observed using a SEM (JIB-PS500i, JEOL, Tokyo, Japan). Finally, the samples were rehydrated in PBS solution for another 24 h before subsequent experiments and observed by a fluorescent microscope (MF52-N, Guangzhou Micro-shot Technology Co., Ltd, Guangzhou, China).
500 mixed in SU-8) was spin coated on the dried samples (1500 rpm, 30 s). After baking (95 °C, 3 min), the samples were exposed to UV irradiation through the designed photomask. Fluorescent SU-8 microdots on the dried PAA gel were obtained after the development process and observed using a SEM (JIB-PS500i, JEOL, Tokyo, Japan). Finally, the samples were rehydrated in PBS solution for another 24 h before subsequent experiments and observed by a fluorescent microscope (MF52-N, Guangzhou Micro-shot Technology Co., Ltd, Guangzhou, China).
The area (S), perimeter (l), Feret's diameter (ddiameter), spacing distance (dspacing) and thickness (dthickness) of SU-8 microdots on PAA gel were measured from SEM and fluorescent images using ImageJ software to characterize the topography of photolithographic microdots. The circularity (γ) of each microdot was calculated from the following eqn (1):
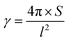 | (1) |
The Young's modulus of the samples in the PAA gel region was characterized through nanoindentation experiments using an AFM (MFP-3D, Oxford Instruments Plc. Abingdon, Oxfordshire, UK) according to previous research.30 The nanoindentation experiments were performed using a silicon nitride cantilever (Novascan, Ames) with a silica sphere (d = 600 nm, k = 0.06 N m−1) and the exact spring constant of each cantilever was verified before measurements. The position of the reference-free TFM and cantilever was observed using a phase-contrast microscope. The rehydrated reference-free TFM was characterized in nanoindentation experiment. During nanoindentation, a 2 nN trigger force and a 4 μm s−1 indentation rate were set. The Poisson ratio was set as 0.5. Young's modulus (E) of reference-free TFM was calculated by fitting the obtained force–distance curves to a Hertz contact model. For each sample, 20 force–distance curves from each position were measured and analyzed. The average Young's modulus (E) from 3 samples was calculated. Photoluminescence (PL) spectroscopy of the photoresist mixture was measured using an imaging spectrometer (iHR 320, HORIBA Ltd, Kyoto, Japan) at a 365 nm excitation wavelength.
Adhesive protein modification of reference-free traction force microscopy
The PAA gel surface of the reference-free TFM was modified by fibronectin, poly-L-lysine, and collagen type I according to a previous report.31 Briefly, the rehydrated reference-free TFM was coated with 200 μL Sulfo-SANPAH (Shanghai Aladdin Biochemical Technology Co., Ltd, Shanghai, China) and further activated by UV irradiation. After rinsing with HEPES solution twice, 20 mg L−1 fibronectin (FN, Beijing Solarbio Science & Technology Co., Ltd, Beijing, China), poly-L-lysine (PLL, Shanghai Aladdin Biochemical Technology Co., Ltd, Shanghai, China) and collagen type I (COL(I), Shanghai Macklin Biochemical Technology Co., Ltd, Shanghai, China) were respectively incubated with the activated reference-free TFM for 24 h at 4 °C. Then, the samples were rinsed with PBS solution twice and placed into 12-well plates before cell culture.Cell culture
In this study, human osteosarcoma cells (MG-63, Procell Life Science & Technology Co., Ltd, Wuhan, China) were selected to investigate the influence of adhesive proteins on cellular contractility. The MG-63 cells were subcultured in DMEM medium (Mishu (Xi’an) Biotechnology Co., Ltd) supplied with 10% FBS (Biological Industries Israel Beit Haemek Ltd) and 1% penicillin–streptomycin (Mishu (Xi’an) Biotechnology Co., Ltd). A 2 mL cell suspension (5000 cells per mL) was added into each well of the TFM and placed in a 12-well plate. After cell culture for 24 h in a humidified CO2 incubator, samples were directly observed by a fluorescent microscope (MF52-N, Guangzhou Micro-shot Technology Co., Ltd, Guangzhou, China).Quantification of cellular contractility
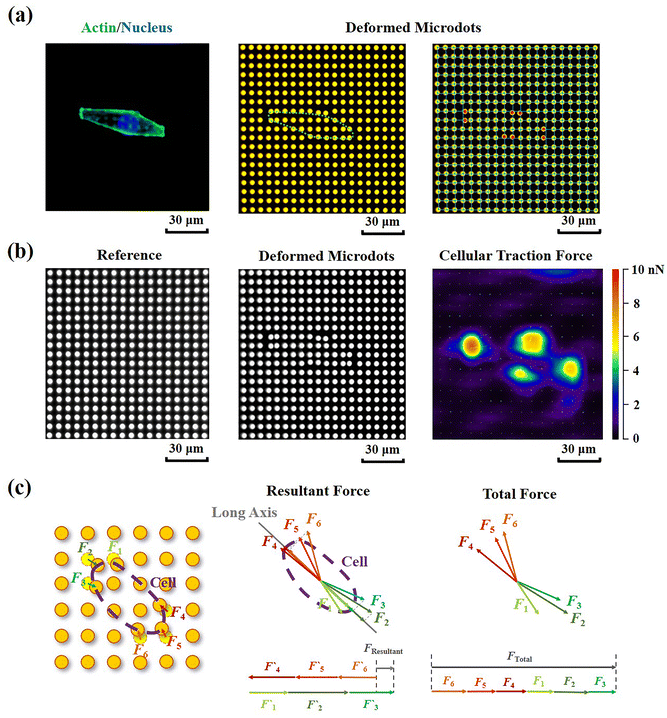
Contractile force was calculated through measurements of the microdot displacement. After cell culture on the reference-free TFM for 24 h, samples were fixed and stained by Actin-Tracker Green-488 (Beyotime Biotechnology Inc., Shanghai, China) and DAPI (Beyotime Biotechnology Inc., Shanghai, China) to obtain the accurate position of adhered cells. Fluorescent images of microdots at the cell adhered region were observed and recorded by a fluorescent microscope (MF52-N, Guangzhou Micro-shot Technology Co., Ltd, Guangzhou, China). The displacement of microdots was analyzed using the MBI-Pillar Tracker plugin in ImageJ software through fast Fourier transform traction (FTT) according to previous report.32 In the prepared reference-free TFM, thin SU-8 microdots were fabricated on the surface of the PAA gel. In this case, the displacement of microdots was related to the deformation of the PAA gel. As shown in Fig. 1d, the PAA gel surface was divided into numerous micro-squares with 6 μm in length. The displacement of each microdot was regarded as the deformation of this micro-squared PAA gel. Additionally, the mechanical property of PAA gel was characterized by nanoindentation as previously described. The result showed a linear relationship between force and indentation depth (Fig. S1, ESI†). Therefore, the cellular contractile force was calculated using eqn (2):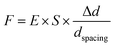 | (2) |
Additionally, in order to characterize the traction state of each cell, the resultant force and total force were calculated, respectively. As shown in Fig. 3c, the resultant force was defined as the vectorial sum of all the traction forces in the direction of the long axis in each cell. While the total force was defined as the summation of the absolute values of traction forces. The distance correlations of scatter plots containing contractile force and cellular morphology were analyzed in this study to determine the relationship between contractile force and cell morphology. The distance correlation coefficient R was calculated by Matlab software according previous report.33R = 0.5 was considered as the threshold for a good correlation. More than 30 cells from 3 independent experiments were analyzed.
Characterization of cell morphology, focal adhesions and actin fibers
The nuclei, actin, and vinculin were stained to characterize the morphological cues and focal adhesion structure of cells on each chemically modified surface. After cells were seeded on chemically modified PAA gels and further incubated under 5% CO2 and 37 °C for 24 h, the cytoskeleton structures were fluorescently stained according to a previous report.34 Briefly, the samples were firstly fixed and permeabilized. Then, an aqueous solution of rabbit anti-vinculin antibody (Abcam plc., Cambridge, UK) at a dilution ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in 2% BSA (Shanghai Aladdin Biochemical Technology Co., Ltd, Shanghai, China) was incubated with the samples at 37 °C for 1 h and further incubated with an aqueous solution of Alexa Fluor-488 labelled goat anti-rabbit IgG antibody (Abcam plc., Cambridge, UK) at a dilution ratio of 1
100 in 2% BSA (Shanghai Aladdin Biochemical Technology Co., Ltd, Shanghai, China) was incubated with the samples at 37 °C for 1 h and further incubated with an aqueous solution of Alexa Fluor-488 labelled goat anti-rabbit IgG antibody (Abcam plc., Cambridge, UK) at a dilution ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 under 37 °C for another 1 h. At last, actin filaments were stained by incubating the samples with Alexa Fluor-594 phalloidin (Beijing Solarbio Science & Technology Co., Ltd, Beijing, China) at a dilution ratio of 1
1000 under 37 °C for another 1 h. At last, actin filaments were stained by incubating the samples with Alexa Fluor-594 phalloidin (Beijing Solarbio Science & Technology Co., Ltd, Beijing, China) at a dilution ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 in PBS at room temperature for 20 min. Nuclei were stained with 10 mg mL−1 of DAPI (Beyotime Biotechnology Inc., Shanghai, China) at room temperature in the dark for 1 h. Finally, the fluorescently stained cytoskeleton structures were observed and recorded by a fluorescent microscope (MF52-N, Guangzhou Micro-shot Technology Co., Ltd, Guangzhou, China).
40 in PBS at room temperature for 20 min. Nuclei were stained with 10 mg mL−1 of DAPI (Beyotime Biotechnology Inc., Shanghai, China) at room temperature in the dark for 1 h. Finally, the fluorescently stained cytoskeleton structures were observed and recorded by a fluorescent microscope (MF52-N, Guangzhou Micro-shot Technology Co., Ltd, Guangzhou, China).
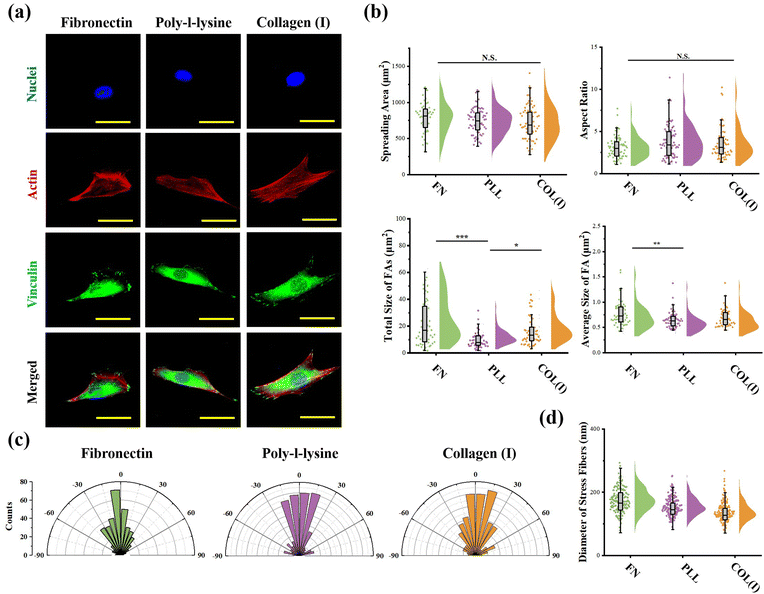
Cellular morphogenesis, including spreading and elongation, was characterized through the analysis of actin and nuclei stained fluorescent images using ImageJ software. The fluorescently stained vinculin was analyzed to characterize the structure of focal adhesions (FAs). The total area and average size of FAs were calculated according to a previous report.35 More than 50 cells from 3 independent experiments were analyzed to obtain quantitative data. The orientation and thickness of stress fibers were measured from actin stained fluorescent images by ImageJ software. For orientation analysis, the orientation of cell's long axis was set as 0 degree. More than 10 cells from each group were analyzed.
Characterization of YAP localization
After osteosarcoma cells were cultured on protein modified surfaces for 24 h, YAP and nuclei were stained to characterize YAP localization according to a previous report.36 Firstly, the samples were fixed with 4% cold paraformaldehyde and permeabilized with 1% Triton X-100. Then, the samples were incubated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 diluted rabbit anti-YAP antibody (Proteintech Group, Inc., Planegg-Martinsried, Germany) for 1 h at 37 °C and subsequently incubated with Alexa Fluor-488 labeled donkey anti-rabbit IgG antibody (Abcam plc., Cambridge, UK) at a dilution ratio of 1
100 diluted rabbit anti-YAP antibody (Proteintech Group, Inc., Planegg-Martinsried, Germany) for 1 h at 37 °C and subsequently incubated with Alexa Fluor-488 labeled donkey anti-rabbit IgG antibody (Abcam plc., Cambridge, UK) at a dilution ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
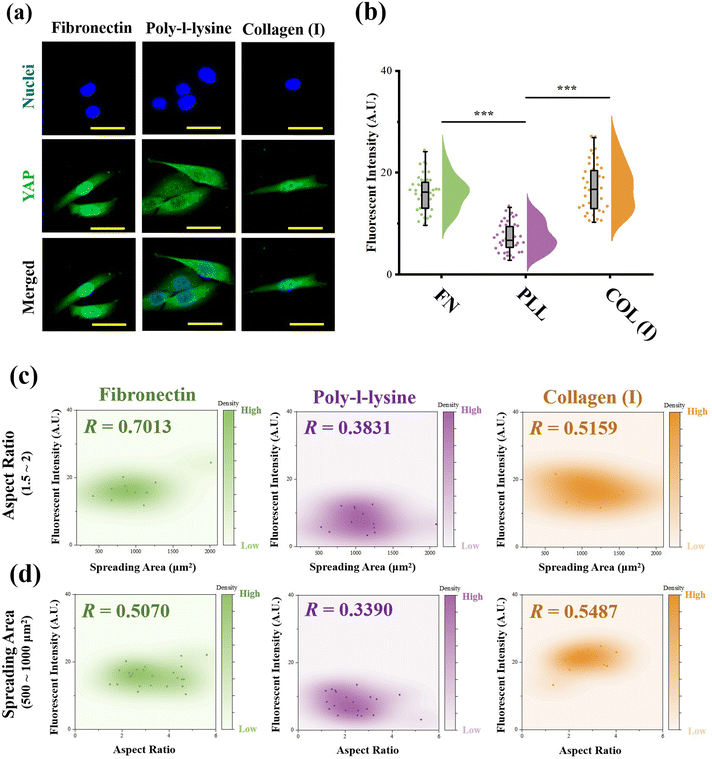
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 for another 1 h at room temperature. Finally, the nuclei were stained with DAPI (Beyotime Biotechnology Inc., Shanghai, China). The fluorescent microscope images were recorded using a laser confocal scanning microscope (LSM800, Carl Zeiss AG, Oberkochen, Germany). The fluorescence intensity of YAP stained cells at the nuclear region was measured from fluorescent images using ImageJ software. More than 30 cells from 3 independent experiments were analyzed to obtain quantitative data. To explore the relationship between YAP activity and cell morphology, scatterplots between fluorescence intensity and cell spreading area or aspect ratio were also analysed by distance correlation, and the coefficient R was also calculated. Similar to the analysis procedure for contractile force, R = 0.5 was also set as the threshold for a good correlation.
1000 for another 1 h at room temperature. Finally, the nuclei were stained with DAPI (Beyotime Biotechnology Inc., Shanghai, China). The fluorescent microscope images were recorded using a laser confocal scanning microscope (LSM800, Carl Zeiss AG, Oberkochen, Germany). The fluorescence intensity of YAP stained cells at the nuclear region was measured from fluorescent images using ImageJ software. More than 30 cells from 3 independent experiments were analyzed to obtain quantitative data. To explore the relationship between YAP activity and cell morphology, scatterplots between fluorescence intensity and cell spreading area or aspect ratio were also analysed by distance correlation, and the coefficient R was also calculated. Similar to the analysis procedure for contractile force, R = 0.5 was also set as the threshold for a good correlation.
Statistical analysis
Statistical analysis was performed using one-way analysis of variance (ANOVA) with Tukey's post hoc test for multiple comparisons to confirm significant differences among samples. The data are presented as means ± standard deviations (SDs). It was considered to be a statistically significant difference when p < 0.05.Results
Measurement of traction force using the prepared reference-free TFM
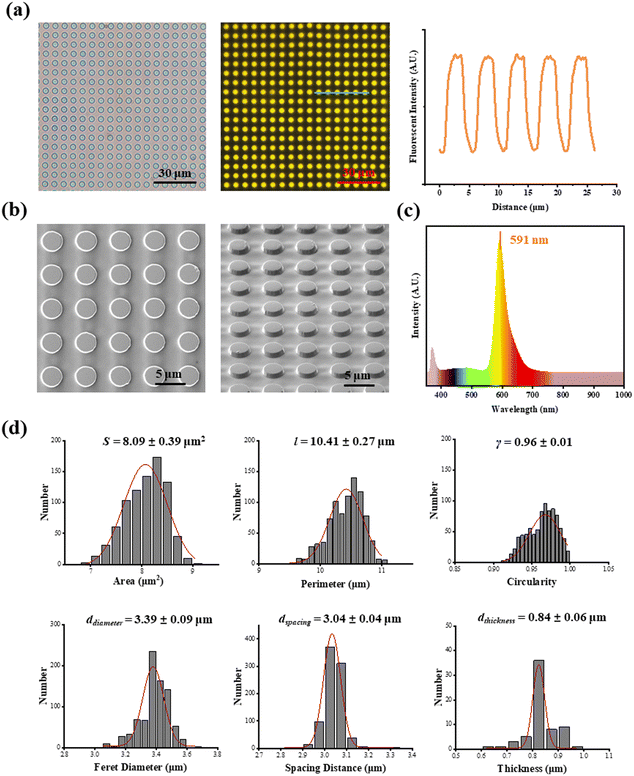
As shown in Fig. 1a, fluorescent SU-8 microdots were designed and fabricated to characterize cellular contractility through the detection of microdot displacement. The microdots were designed as 3 μm in diameter and 3 μm in spacing (Fig. 1b). Initially, a well-organized array of these microdots was prepared on the surface of elastic PAA gel. Subsequently, the PAA gel surfaces were chemically modified with adhesive proteins, including fibronectin (FN), poly-L-lysine (PLL), and collagen type I (COL(I)). Following cell adhesion, the contraction-induced displacement of SU-8 microdots led to the deformation of the PAA gel, allowing measurement of cellular contractility by characterizing of microdot displacement.The prepared reference-free TFM showed a well-organized microdot array in both phase-contrast and fluorescent microscope images (Fig. 2a). The distinct yellow fluorescent points in Fig. 2a confirmed the suitability of the array for detecting the microdot displacement. SEM images further revealed the regular structures of the microdots’ array on the surface of PAA gel (Fig. 2b). According to the results, the area, perimeter, Feret's diameter, spacing and circularity of the microdots were 8.09 ± 0.39 μm2, 10.41 ± 0.27 μm, 3.39 ± 0.09 μm, 3.04 ± 0.04 μm, and 0.96 ± 0.01, respectively (Fig. 2d). In addition, the Young's modulus of the PAA gel measured from nanoindentation experiments was 2.77 ± 0.13 kPa. These results confirmed successful preparation of the designed microdot array on PAA gel surface. Furthermore, the thickness of SU-8 microdots was less than 1 μm (0.84 ± 0.06 μm), ensuring minimal impact on the elasticity of the PAA gel.
Cellular contractility was measured through the detection of microdot displacement once the cells were cultured on the TFM. As shown in Fig. 3a, actin and nuclei were firstly stained to confirm the cell adhered region, and the fluorescent images of microdots were observed and recorded to analyze cell contractility. As shown in the right row of Fig. 3a, microdots obviously moved in regions where cells adhered, indicating contractile cell adhesion. These displaced microdots were directly observed and recorded by a traditional fluorescent microscope. Compared with the reference image without cell adhesion (Fig. 3b), the displacement of microdots can be easily analyzed to calculate cellular contractility according to eqn (2). It was worth noting that, benefitted from the ability of noise decreasing and nonlinear elasticity correction, numerous mathematical solutions including finite elements analysis (FEA),27 Fourier transform traction cytometry (FTTC),32 regularized Fourier transform traction cytometry (reg-FTTC)37 and integral Boussinesq solution38 were suggested to be more accurate in calculation of traction force. These mathematical methods may also be suitable for the analysis of microdots’ displacement images from designed reference-free TFM. Given that the resultant force and total force of contractility have been demonstrated to be related to cellular morphogenesis and migration,39 both the resultant force and total force of contractile cells were calculated, and their relationship with cellular morphology was analyzed (Fig. 3c).
Effects of adhesive proteins on cellular contractility
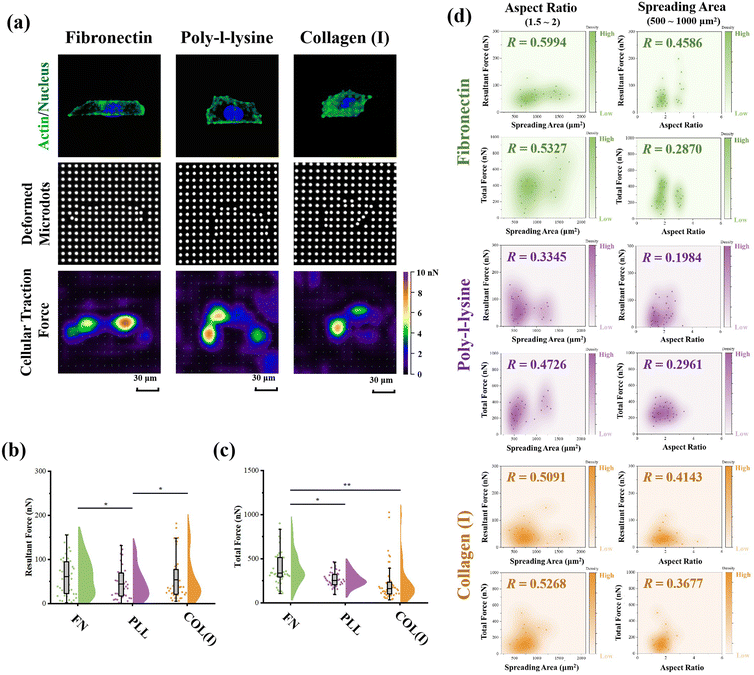
The PAA gel surfaces in the reference-free TFM were chemically modified by FN, PLL and COL(I) to investigate the effects of adhesive proteins on cellular contractility. In the results, cells successfully adhered on each chemically modified reference-free TFM surfaces, displaying a well-spread morphology (Fig. 4a). After calculation of the resultant force and total force of contractile cells, the effects of ECM components on cellular contractility were characterized. The resultant force of cells adhered on PLL-modified surfaces was significantly lower than that of cells adhered on FN- or COL(I)-modified surfaces (Fig. 4b). Additionally, the total force of cells adhered on PLL-modified surfaces was lower than that of cells adhered on FN-modified surfaces. These results indicated the weak effect of PLL on the induction of cellular contractility.In addition, the relationship between cellular contractility and cell morphology was further analyzed. As shown in Fig. S2 (ESI†), both the resultant force and total force were independent of cell spreading and elongation (R < 0.5). Otherwise, cellular morphology was tightly related to cell contractility40 and cells adhered on TFM surfaces showed a variety of morphologies. In this case, only the cells with a similar spreading area or aspect ratio were analyzed to obtain more reasonable results. In particular, cells with 500 to 1000 μm2 in spreading area were selected to analyze the relationship between cell elongation and cellular contractility. Simultaneously, cells with an aspect ratio from 1.5 to 2 were selected to analyze the correlation of cell spreading and cellular contractility. As shown in Fig. 4d, on FN- or COL(I)-modified surfaces, both the resultant force and total force of contractile cells were obviously positively related to the cell spreading area (R > 0.5). On the other hand, when cells adhered on PLL-modified surfaces, both resultant force and total force were revealed to be independent of cell spreading (R < 0.5). These results indicated that both FN and COL(I) in ECM were able to break the steady state of force and improve cell contractility with cell spreading but PLL was inefficient at regulating of cellular contractility. Additionally, the relationship between cellular contractility and cell elongation was also analyzed. In the results, the efficacy of cellular elongation in regulation of contractile force was weaker than cell spreading (R < 0.5). In addition, for the cells cultured on FN- or COL(I)-modified surfaces, cell elongation was more efficient at regulating of resultant force (R = 0.4586 and 0.4143) than total force (R = 0.2870 and 0.3677). Nevertheless, the tighter correlation between cell elongation and contractile force for cells adhered on FN- and COL(I)-modified surfaces also revealed the potential effects of this morphogenesis on cellular contractility was stronger than the cells adhered on PLL-modified surfaces.
Effects of adhesive proteins on cellular morphogenesis, FA structures and stress fibers
The effects of adhesive proteins on contractile cell morphogenesis and FA structures were characterized. As shown in Fig. 5a, the nuclei, actin, and vinculin of contractile cells cultured on each chemically modified surface were stained. In the results, adhesive proteins were inefficient in the regulation of cell spreading and elongation (Fig. 5b). Moreover, adhesive proteins showed a significant influence on the size of FAs. Specifically, both the total amount and size of FAs assembled in cells adhered on PLL-modified surfaces were significantly lower than in cells adhered on FN- or COL(I)-modified surfaces. These results indicated that compared with FN or COL(I), even though PLL was able to provide a suitable environment for cell spreading and elongation, it was inefficient in the promotion of FA assembly. Additionally, orientation and thickness of stress fibers were also analyzed. In the results, most of the stress fibers were orientated along the long axis of the cell. In particular, more unified orientation was observed in cells adhered on FN-modified surfaces (Fig. 5c). Simultaneously, the thickness of stress fibers showed no significant difference between each adhesive protein modified surfaces (Fig. 5d).Effects of adhesive proteins on YAP activation
As the most critical mechanosensitive transcription factor, the localization of YAP has always been considered as the benchmark to indicate YAP activity.41 Therefore, the fluorescence intensity of stained YAP at the nuclei region was measured (Fig. 6a). In the results, cells adhered on PLL-modified surfaces showed significantly lower fluorescence intensity at the nuclear region than cells cultured on FN- or COL(I)-modified surfaces. In addition, the relationship between YAP localization and cell morphology was analyzed. Similar to the results of the relationship between cellular contractility and cell morphology, YAP activity showed independence with cell elongation and spreading (R < 0.5, Fig. S3, ESI†). Therefore, the cells with similar spreading area or aspect ratio were also selected and analyzed. As shown in Fig. 6c and d, when cells were cultured on FN- or COL(I)- modified surfaces, YAP activity was positively related to cell spreading and elongation (R < 0.5) Compared with the tight relationship between cell morphology and YAP activity on FN-modified surfaces (R = 0.7013 and 0.5070), the correlation was weaker when cells adhered on COL(I)-modified surfaces (R = 0.5159 and 0.5487). It may be caused by the activation of YAP by COL(I).42 It may also be the reason for the highest YAP activity on COL(I)-modified surfaces. Different from cells cultured on FN or COL(I), YAP activity was independent of cell spreading (R = 0.3831) and elongation (R = 0.3890) on PLL-modified surfaces. These results revealed that cell spreading and elongation can activate YAP when cells adhered to FN- or COL(I)-modified surfaces. In contrast, cell spreading and elongation were independent of YAP activity when cells were cultured on PLL-modified surfaces.Discussion
In the present research, a reference-free TFM was simply prepared through photolithography using approachable reagents. In traditional TFM with randomly distributed fluorescent dots, in situ observation of attached cell exfoliation was necessary and made it laborious toin observe numerous samples. In these researches, a typical image with the distribution of contractile force was commonly shown but difficult to obtain statistical results.43 Benefited from the well-organized SU-8 array structures, a reference image was unnecessary in the detection of microdot displacement and more than 50 cells from each group were analyzed to sufficient samples. In addition, the designed SU-8 array could be observed through a traditional fluorescent microscope. With these advantages, the prepared reference-free TFM provides a high-throughput method for the measurement of cellular contractility. By using the prepared reference-free TFM, the influence of adhesive proteins in the ECM on cellular contractility, cell morphology, and mechanotransduction was investigated.In summary, an obviously higher contractility was observed when cells adhered on FN-/COL(I)- modified surfaces (Fig. 4b and c). Interestingly, there was another research showed the increased density of COL(I) was able to enhance cellular contractility.44 Even the excessed COL(I) was able to limits the contraction, but it was only caused by the adhesive substrate.45 These results revealed the critical role of COL(I) in regulation of cellular contractility. Additionally, fibronectin was also demonstrated to be essential in the regulation of cellular contractility through fibronectin polymerization.46 Based on previous reports, cellular contractility originates from the motion of the molecular motor myosin II on actin filaments.47 These contractile filaments are linked with integrins and further adhere to adhesive proteins in the ECM.48 In addition, as an integrin-containing multi protein structure,49 focal adhesions are always considered as a mechanical link between the ECM and cytoskeleton,50 further influencing on various cell behaviors through mechanotransduction.51 Among adhesive proteins, FN is able to connect with integrin receptors on the cell membrane and further bind with the actin cytoskeleton.52 Similar to FN, cells adhered on COL(I) are also associated with integrin.53 As shown in Fig. 5b, more and larger FAs assembled in cells cultured on FN- or COL(I)-modified surfaces with integrin associated adhesion. Consequently, as shown in Fig. 7 the asymmetric contractile force and promoted contractility on FN- or COL(I)-modified surfaces (Fig. 4b and c) were considered to be the results of denser integrin associated cell adhesion. Moreover, previous reports indicated that contractile force was able to induce cellular morphogenesis.54 Simultaneously, some other research studies also demonstrated that cell morphology was able to regulate the state of cell contractility by using morphology confined cells.55,56 All of these researches revealed the tight relationship between cellular contractility and morphogenesis. With a comprehensive analysis of the results shown in Fig. 4d, this tight relationship only occurred with integrin associated adhesion. There was another interesting phenomenon that the total force of cells adhered on COL(I)-modified surfaces was significantly lower than the cells adhered on FN-modified surfaces (Fig. 4c). Since the total force was tightly related to vinculin associated cell adhesions, the lower total force of COL(I)- may be caused by the slight smaller FAs and less unified orientation of stress fibers (Fig. 5b and c). Furthermore, the resultant force was correlated to both cell spreading and elongation but the total force was only related to cell spreading in cells adhered on FN- or COL(I)-modified surfaces (Fig. 4d). Based on previous report, the resultant force was always related to asymmetric morphogenesis.57 Additionally, cell spreading and elongation was tightly related to mitosis and migration which were always accompanied with asymmetric morphogenesis.30,58,59 This was considered as the reason of spreading and elongation related resultant force. Furthermore, the total contractile force was demonstrated to be related to adhesion strength60 and the detachment process of metastasis in vinculin associated cell adhesion.61 These biological processes were always associated with cell spreding.62,63 In this case, the total force was only related to cell spreading.
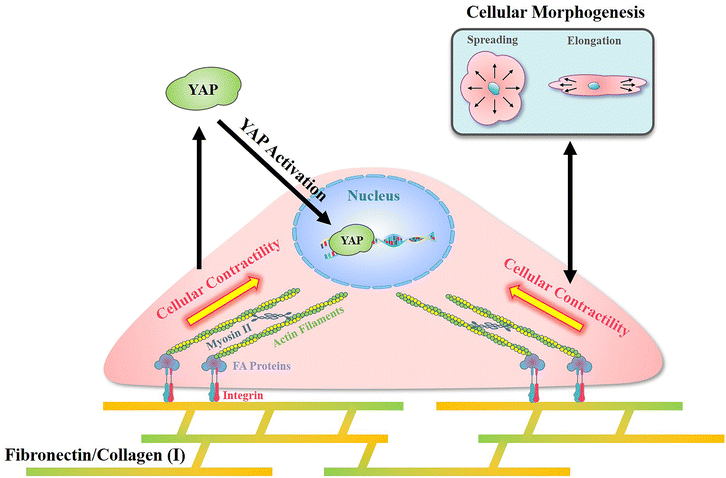 | ||
| Fig. 7 Illustration of the mechanism of cellular contractility induced morphogenesis and YAP activation with integrin associated cell adhesion on fibronectin and collagen type I. | ||
In addition, with integrin associated cell adhesion, YAP activity was related to cell morphology (Fig. 6c and d). These results were consistent with previous reports that cell morphogenesis was able to regulate YAP activity.64 On the other hand, some research studies demonstrated cellular contractility was also able to regulate YAP activity.65 Since cellular morphogenesis always processed with changing of cellular contractility when cells were integrin associated adhesion, the different roles of them in the regulation of YAP activity remains unclear. Nevertheless, as shown in Fig. 7, cell morphology and contractility were considered to be effective regulators of YAP activity in cells cultured on FN or COL(I) with integrin associated adhesions.
Compared with FN or COL(I), PLL is a kind of cationic biopolymer that is considered as a nonspecific attachment macromolecule for cell adhesion. Typically, the cationic PLL is able to interact with polyanionic plasma membrane and has been widely applied to promote cell adhesion.66 In the results, even though PLL was able to support cell spreading and elongation, it was insufficient for the promotion of FA assembly (Fig. 5b) and cellular contractility (Fig. 4b and c). Furthermore, cellular contractility and YAP activity were independent of cellular morphogenesis when cells were cultured on PLL modified surfaces (Fig. 4d and 6c). These results illustrated that without integrin associated adhesion, both cellular contractility and YAP were unable to be activated. Additionally, it is worth noting that by using PLL modified surfaces, cellular contractility was blocked without affecting cellular morphogenesis. Based on the results of decreased YAP activity, the predominant role of cellular contractility in the regulation of mechanotransduction was revealed.
Conclusions
In summary, a reference-free TFM was prepared to detect cellular contractility, and the effects of adhesive proteins on cellular contractility were investigated using the prepared TFM in this study. The results, showed that both fibronectin and collagen(I) were able to promote cellular contractility and YAP activity through integrin associated adhesion. In contrast, even poly-L-lysine was able to support cellular spreading and elongation but was inefficient for the activation of cellular contractility and mechanotransduction. In addition, compared with cell morphology, YAP activity was predominantly regulated by cellular contractility.Author contributions
Conceptualization: Y. Y. and X. W.; funding acquisition: Y. Y. and B. X.; investigation: Y. Y., K. H. and Y. W.; methodology, all co-authors; project administration, Y. Y. and S. W.; resources, Y. Y., Y. W. and X. W.; data curation, all co-authors; supervision: Y. Y. and X. W.; visualization, Y. Y., S. H. and Y. W.; writing – original draft preparation: Y. Y., S. D., S. W, Y. W and X. W.; and writing – review and editing: all co-authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant no. 32201124 and 82302401), the Foundation of Shaanxi University of Science and Technology (Grant no. 126021993), Shanxi-Zheda Institute of Advanced Materials and Chemical Engineering, Shanghai Overseas High-Level Talent Project and High-End Foreign Experts Introduction Plan of Ministry of Science and Technology (No. G2023013020L).References
- G. M. Walker, H. C. Zeringue and D. J. Beebe, Lab Chip, 2004, 4, 91–97 RSC.
- G. Nardone, J. Oliver-De La Cruz, J. Vrbsky, C. Martini, J. Pribyl, P. Skladal, M. Pesl, G. Caluori, S. Pagliari, F. Martino, Z. Maceckova, M. Hajduch, A. Sanz-Garcia, N. M. Pugno, G. B. Stokin and G. Forte, Nat. Commun., 2017, 8, 15321 CrossRef CAS PubMed.
- Y. Yang, X. Wang, Y. Wang, X. Hu, N. Kawazoe, Y. Yang and G. Chen, Sci. Rep., 2019, 9, 6891 CrossRef PubMed.
- N. Wang, J. P. Butler and D. E. Ingber, Science, 1993, 260, 1124–1127 CrossRef CAS PubMed.
- V. S. Deshpande, R. M. McMeeking and A. G. Evans, Proc. R. Soc. A, 2007, 463, 787–815 CrossRef CAS.
- E. G. Rens and R. M. H. Merks, Biophys. J., 2017, 112, 755–766 CrossRef CAS PubMed.
- S.-J. Heo, Woojin M. Han, Spencer E. Szczesny, Brian D. Cosgrove, Dawn M. Elliott, David A. Lee, Randall L. Duncan and Robert L. Mauck, Biophys. J., 2016, 111, 864–874 CrossRef CAS PubMed.
- R. G. Thakar, M. G. Chown, A. Patel, L. Peng, S. Kumar and T. A. Desai, Small, 2008, 4, 1416–1424 CrossRef CAS PubMed.
- P. Pandya, J. L. Orgaz and V. Sanz-Moreno, Curr. Opin. Cell Biol., 2017, 48, 87–96 CrossRef CAS PubMed.
- E. K. F. Yim and M. P. Sheetz, Stem Cell Res. Ther., 2012, 3, 41 CrossRef CAS PubMed.
- M. A. Wozniak and C. S. Chen, Nat. Rev. Mol. Cell Biol., 2009, 10, 34–43 CrossRef CAS PubMed.
- J. P. Hoj, J. A. Davis, K. E. Fullmer, D. J. Morrell, N. E. Saguibo, J. T. Schuler, K. J. Tuttle and M. D. H. Hansen, Exp. Cell Res., 2014, 326, 187–200 CrossRef CAS PubMed.
- T. Wang, S. Hamilla, M. Cam, H. Aranda-Espinoza and S. Mili, Nat. Commun., 2017, 8, 896 CrossRef PubMed.
- R. G. Thakar, M. G. Chown, A. Patel, L. Peng, S. Kumar and T. A. Desai, Small, 2008, 4, 1416–1424 CrossRef CAS PubMed.
- S. Schlie-Wolter, A. Ngezahayo and B. N. Chichkov, Exp. Cell Res., 2013, 319, 1553–1561 CrossRef CAS PubMed.
- A. Katsumi, A. W. Orr, E. Tzima and M. A. Schwartz, J. Biol. Chem., 2004, 279, 12001–12004 CrossRef CAS PubMed.
- D. E. Leckband and J. de Rooij, Annu. Rev. Cell Dev. Biol., 2014, 30, 291–315 CrossRef CAS PubMed.
- T. D. Ross, B. G. Coon, S. Yun, N. Baeyens, K. Tanaka, M. Ouyang and M. A. Schwartz, Curr. Opin. Cell Biol., 2013, 25, 613–618 CrossRef CAS PubMed.
- A. R. Chethikkattuveli Salih, K. Hyun, A. Asif, A. M. Soomro, H. M. U. Farooqi, Y. S. Kim, K. H. Kim, J. W. Lee, D. Huh and K. H. Choi, Polymers, 2021, 13, 3016 CrossRef CAS PubMed.
- P. J. Albert and U. S. Schwarz, Cell Adhes. Migr., 2016, 10, 516–528 CrossRef CAS PubMed.
- W. Ronan, V. S. Deshpande, R. M. McMeeking and J. P. McGarry, Biomech. Model. Mechanobiol., 2014, 13, 417–435 CrossRef PubMed.
- A. K. Harris, P. Wild and D. Stopak, Science, 1980, 208, 177–179 CrossRef CAS PubMed.
- H. Colin-York and M. Fritzsche, Curr. Opin. Biomed. Eng., 2018, 5, 1–5 CrossRef.
- J. L. Teo, C. T. Lim, A. S. Yap and T. B. Saw, STAR Protoc., 2020, 1, 100098 CrossRef PubMed.
- J. H. C. Wang and B. Li, in Cytoskeleton Methods and Protocols, ed. R. H. Gavin, Humana Press, Totowa, NJ, 2010, pp. 301–313 DOI:10.1007/978-1-60761-376-3_17.
- O. A. Banda, C. R. Sabanayagam and J. H. Slater, ACS Appl. Mater. Interfaces, 2019, 11, 18233–18241 CrossRef CAS PubMed.
- M. Bergert, T. Lendenmann, M. Zündel, A. E. Ehret, D. Panozzo, P. Richner, D. K. Kim, S. J. P. Kress, D. J. Norris, O. Sorkine-Hornung, E. Mazza, D. Poulikakos and A. Ferrari, Nat. Commun., 2016, 7, 12814 CrossRef CAS PubMed.
- I. Schoen, W. Hu, E. Klotzsch and V. Vogel, Nano Lett., 2010, 10, 1823–1830 CrossRef CAS PubMed.
- K. M. Beussman, M. Y. Mollica, A. Leonard, J. Miles, J. Hocter, Z. Song, M. Stolla, S. J. Han, A. Emery, W. E. Thomas and N. J. Sniadecki, Acta Biomater., 2023, 163, 302–311 CrossRef CAS PubMed.
- Y. Yang, X. Wang, X. Hu, N. Kawazoe, Y. Yang and G. Chen, ACS Appl. Mater. Interfaces, 2019, 11, 1932–1941 CrossRef CAS PubMed.
- M. Dembo and Y.-L. Wang, Biophys. J., 1999, 76, 2307–2316 CrossRef CAS PubMed.
- J. P. Butler, I. M. Tolić-Nørrelykke, B. Fabry and J. J. Fredberg, Am. J. Physiol.: Cell Physiol., 2002, 282, C595–C605 CrossRef CAS PubMed.
- A. Bhattacharjee, J. Modern Appl. Statistical Methods, 2014, 13(1), 354–366 CrossRef.
- Y. Yang, X. Wang, T. C. Huang, X. Hu, N. Kawazoe, W. B. Tsai, Y. Yang and G. Chen, J. Mater. Chem. B, 2018, 6, 5424–5434 RSC.
- S. Huang, Z. Chen, X. Hou, K. Han, B. Xu, M. Zhang, S. Ding, Y. Wang and Y. Yang, Int. J. Mol. Sci., 2022, 23(19), 11884 CrossRef CAS PubMed.
- Y. Wang, Y. Yang, X. Wang, N. Kawazoe, Y. Yang and G. Chen, Acta Biomater., 2021, 125, 100–111 CrossRef CAS PubMed.
- A. H. Kulkarni, P. Ghosh, A. Seetharaman, P. Kondaiah and N. Gundiah, Soft Matter, 2018, 14, 4687–4695 RSC.
- J. Huang, X. Peng, L. Qin, T. Zhu, C. Xiong, Y. Zhang and J. Fang, J. Biomech. Eng., 2009, 131, 061009 CrossRef PubMed.
- M. Murrell, P. W. Oakes, M. Lenz and M. L. Gardel, Nat. Rev. Mol. Cell Biol., 2015, 16, 486–498 CrossRef CAS PubMed.
- I. B. Bischofs, F. Klein, D. Lehnert, M. Bastmeyer and U. S. Schwarz, Biophys. J., 2008, 95, 3488–3496 CrossRef CAS PubMed.
- A. Elosegui-Artola, I. Andreu, A. E. M. Beedle, A. Lezamiz, M. Uroz, A. J. Kosmalska, R. Oria, J. Z. Kechagia, P. Rico-Lastres, A.-L. Le Roux, C. M. Shanahan, X. Trepat, D. Navajas, S. Garcia-Manyes and P. Roca-Cusachs, Cell, 2017, 171(1397–1410), e1314 Search PubMed.
- Q. Xu, X. Liu, W. Liu, T. Hayashi, M. Yamato, H. Fujisaki, S. Hattori, S. I. Tashiro, S. Onodera and T. Ikejima, Mol. Cell. Biochem., 2019, 450, 87–96 CrossRef CAS PubMed.
- R. W. Style, R. Boltyanskiy, G. K. German, C. Hyland, C. W. MacMinn, A. F. Mertz, L. A. Wilen, Y. Xu and E. R. Dufresne, Soft Matter, 2014, 10, 4047–4055 RSC.
- A. Das, J. Carcinog. Mutagen., 2013, S13, 003 Search PubMed.
- C. Gaudet, W. A. Marganski, S. Kim, C. T. Brown, V. Gunderia, M. Dembo and J. Y. Wong, Biophys. J., 2003, 85, 3329–3335 CrossRef CAS PubMed.
- D. C. Hocking, J. Sottile and K. J. Langenbach, J. Biol. Chem., 2000, 275, 10673–10682 CrossRef CAS PubMed.
- I. Rayment, H. M. Holden, M. Whittaker, C. B. Yohn, M. Lorenz, K. C. Holmes and R. A. Milligan, Science, 1993, 261, 58–65 CrossRef CAS PubMed.
- I. Delon and N. H. Brown, Curr. Opin. Cell Biol., 2007, 19, 43–50 CrossRef CAS PubMed.
- P. Kanchanawong, G. Shtengel, A. M. Pasapera, E. B. Ramko, M. W. Davidson, H. F. Hess and C. M. Waterman, Nature, 2010, 468, 580–584 CrossRef CAS PubMed.
- M. A. Wozniak, K. Modzelewska, L. Kwong and P. J. Keely, Biochim. Biophys. Acta, Mol. Cell Res., 2004, 1692, 103–119 CrossRef CAS PubMed.
- M. A. Schwartz and D. W. DeSimone, Curr. Opin. Cell Biol., 2008, 20, 551–556 CrossRef CAS PubMed.
- C. T. Hsiao, H. W. Cheng, C. M. Huang, H. R. Li, M. H. Ou, J. R. Huang, K. H. Khoo, H. W. Yu, Y. Q. Chen, Y. K. Wang, A. Chiou and J. C. Kuo, Oncotarget, 2017, 8, 70653–70668 CrossRef PubMed.
- J. Jokinen, E. Dadu, P. Nykvist, J. Käpylä, D. J. White, J. Ivaska, P. Vehviläinen, H. Reunanen, H. Larjava, L. Häkkinen and J. Heino, J. Biol. Chem., 2004, 279, 31956–31963 CrossRef CAS PubMed.
- Andrew G. Clark, O. Wartlick, G. Salbreux and Ewa K. Paluch, Curr. Biol., 2014, 24, R484–R494 CrossRef CAS PubMed.
- C. Y. Tay, Y.-L. Wu, P. Cai, N. S. Tan, S. S. Venkatraman, X. Chen and L. P. Tan, NPG Asia Mater., 2015, 7, e199 CrossRef CAS.
- A. Ghagre, A. Amini, L. K. Srivastava, P. Tirgar, A. Khavari, N. Koushki and A. Ehrlicher, ACS Appl. Mater. Interfaces, 2021, 13, 19726–19735 CrossRef CAS PubMed.
- J. L. Maître, H. Turlier, R. Illukkumbura, B. Eismann, R. Niwayama, F. Nédélec and T. Hiiragi, Nature, 2016, 536, 344–348 CrossRef PubMed.
- F. Roegiers and Y. N. Jan, Curr. Opin. Cell Biol., 2004, 16, 195–205 CrossRef CAS PubMed.
- L. Zhang, J. Luo, P. Wan, J. Wu, F. Laski and J. Chen, Development, 2011, 138, 455–464 CrossRef CAS PubMed.
- D. W. Dumbauld, H. Shin, N. D. Gallant, K. E. Michael, H. Radhakrishna and A. J. García, J. Cell. Physiol., 2010, 223, 746–756 CrossRef CAS PubMed.
- N. Paddillaya, K. Ingale, C. Gaikwad, D. K. Saini, P. Pullarkat, P. Kondaiah, G. I. Menon and N. Gundiah, Soft Matter, 2022, 18, 4378–4388 RSC.
- K. K. Elineni and N. D. Gallant, Biophys. J., 2011, 101, 2903–2911 CrossRef CAS PubMed.
- L. Weiss and P. M. Ward, Cancer Metastasis Rev., 1983, 2, 111–127 CrossRef CAS PubMed.
- Y. Wang, N. Wang, Y. Yang, Y. Chen and Z. Zhang, J. Nanobiotechnol., 2022, 20, 499 CrossRef CAS PubMed.
- A. Das, R. S. Fischer, D. Pan and C. M. Waterman, J. Biol. Chem., 2016, 291, 6096–6110 CrossRef CAS PubMed.
- D. Mazia, G. Schatten and W. Sale, J. Cell Biol., 1975, 66, 198–200 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4tb00065j |
| This journal is © The Royal Society of Chemistry 2024 |