Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Advancing nucleic acid delivery through cationic polymer design: non-cationic building blocks from the toolbox
Ilaria
Porello
 ,
Nina
Bono
,
Nina
Bono
 ,
Gabriele
Candiani
,
Gabriele
Candiani
 and
Francesco
Cellesi
and
Francesco
Cellesi
 *
*
Department of Chemistry, Materials and Chemical Engineering “G. Natta”, Politecnico di Milano, Via Luigi Mancinelli 7, 20131, Milan, Italy. E-mail: francesco.cellesi@polimi.it
First published on 24th June 2024
Abstract
Polymers used for the delivery of nucleic acids (NAs) typically possess ionizable, cationic moieties enabling their electrostatic interactions with negatively charged NAs and form stable polyplexes. However, non-cationic building blocks have been harnessed to design cationic polymers with enhanced delivery of DNA/RNA to tissues, cells, and subcellular compartments while remaining stable in biological fluids. By customizing the chemistry of these functional groups, we can improve cell targeting behavior, uptake, endosomal escape, non-toxicity, and transfection efficiency. Additionally, the physicochemical properties, such as the loading capacity, complexation ability, size and morphology, biodegradability, pH sensitivity, and amphiphilicity, can be adjusted based on the specific application. This review summarizes the role of non-cationic moieties in various biomedical contexts, from therapeutic interventions to gene editing. By unpacking and critically summarizing the existing literature, this review provides valuable insights into the rational integration of these building blocks for designing more effective nanovectors to deliver NAs.
1. Introduction
Delivery of nucleic acids (NAs) into mammalian cells represents a ground-breaking advancement in molecular medicine,1 holding significant promise for genetically treating various diseases, including inherited disorders, certain types of cancer,2 and specific viral infections (i.e., COVID-19 vaccines).3–5 Additionally, it is pivotal in basic research because it allows the exploration of cellular mechanisms and the production of specific proteins.6By definition, gene-based therapy is the deliberate modulation of gene expression in target cells accomplished by introducing exogenous NAs such as DNA,7 RNA (mRNA, small or short interfering RNA (siRNA), short hairpin RNA (shRNA),8,9 microRNA (miRNA)),10 antisense oligonucleotides (ASOs),11 and CpG oligodeoxynucleotides or oligoribonucleotides (CpG-ODNs or CpG-ORNs, respectively).12–14 Given the large size and the negative charge of these macromolecules, which limit their ability to freely cross the cell membrane (i.e., at physiological pH they are both anionic, resulting in electrostatic repulsion), and their limited extracellular stability, the intracellular delivery of NAs is typically achieved using physical/mechanical methods15–17 or mediated by vectors.18–21
Physical and mechanical methods, such as electroporation,22 sonoporation,23 magnetofection,24 optoporation,25 gene gun,26 and microinjection,27 drive naked NAs into the cytosol or the cell nucleus to achieve successful gene transfer. Despite their potential and the attention garnered from scientists, the limitations of these technologies make them less attractive than others for NA transfer purposes. Indeed, they often cause toxicity and are not very effective when used in vivo. A straightforward way to deliver NAs into cells relies on the use of gene delivery carriers (vectors), categorized as viral and non-viral. Engineered viral vectors, wherein the therapeutic gene cassette replaces a portion of the viral genome, currently stand as the most extensively utilized carriers in gene therapy, owing to their natural ability to enter host cells to give high transduction efficiency.18,28,29 While significantly advancing the field of gene therapy, viral vectors come also with several drawbacks, including carcinogenesis, immunogenicity, broad tropism, limited DNA packaging capacity, and challenges in vector production.30–33 Non-viral gene delivery (i.e., a process called transfection) holds promise in addressing many of these limitations, especially when it comes to safety. For instance, synthetic vehicles are generally less immunogenic as compared to viral vectors, and patients lack pre-existing immunity, as is the case with certain viral systems. This results in non-viral vectors that are safer and better tolerated by the human body, enabling repeated administration in patients affected by diseases requiring long-term treatment, such as chronic conditions, without causing an immunological response or toxic accumulation in off-target organs. Non-viral vectors also offer the potential to deliver larger genetic payloads and are typically easier to synthesize in a more reproducible manner than their viral counterparts.21,34,35 In most cases, non-viral vectors rely on the use of cationic lipids (CLs),7,20 cationic polymers (CPs),21,36–38 or copolymers based on the combination of cationic monomers with hydrophobic and hydrophilic ones.39–41 These macromolecules self-assemble with anionic NAs to form particle-like complexes (lipoplexes and polyplexes for CLs and CPs, respectively, or micelleplexes for amphiphilic CPs) ranging in hydrodynamic diameter from tens of nanometres to a few micrometres. The small size eases their penetration through the cellular membrane and subsequent NA internalization into cells. Unfortunately, non-viral vectors still possess lower transfection efficiency than their viral counterparts. On the other hand, they can be deliberately designed to encompass multiple functionalities, allowing them to navigate and overcome diverse biological barriers encountered in vitro and in vivo throughout the gene delivery process.
In this context, structure–activity relationship (SAR) studies of non-viral carriers have paved the way for improving their effectiveness and creating customized, functionalized architectures42 with specific physicochemical features that ease intracellular delivery and precise cargo release at the target site.43,44
2. Challenges in nucleic acid delivery
To maximize the effectiveness of non-viral gene delivery carriers, their transfection efficiency must be improved.45,46Regardless of the in vivo administration route (e.g., inhalation, intramuscular injection, intravascular injection), non-viral complexes shall inevitably face the extracellular environments that blunt the fraction of nanovectors that reach the target therapeutic site. The optimal delivery vehicle should be neither cytotoxic nor immunogenic, protect the payload from nuclease degradation during the delivery process, and mediate the intracellular delivery of NA cargo. Further requirements include the control of the pharmacokinetics, distribution, and accumulation at the target site.47
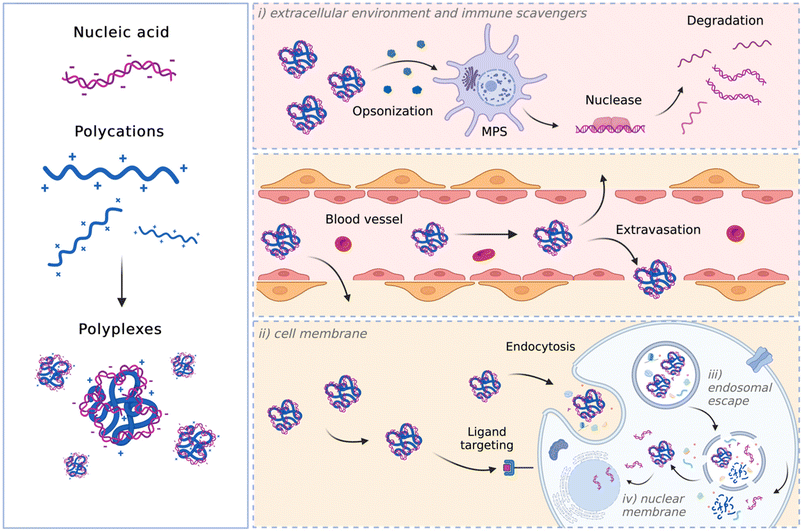
The biological barriers that polyplexes encounter depend on the route of administration.48 Although local delivery may help avoiding some hurdles faced with systemic delivery, the former typically requires deploying more complex techniques that pose other limitations. Furthermore, local delivery may only be useful in diseases where the pathology is restricted to known, accessible sites, such as certain solid cancers or traumatic injuries, so systemic administration is preferred. If limited to systemic administration, a successful carrier shall: (i) have prolonged plasma circulation and be able to cross the vessel wall, (ii) be internalized by cells passing through the plasma membrane, (iii) escape the endosome and release NAs intracellularly, and, in the case of the DNA cargo, (iv) enter the nucleus. A schematic representation of these in vivo hurdles is depicted in Fig. 1.
Within the circulatory system, various determinants, including serum proteins and blood cells, can hinder the biological activity of the particles, leading to their rapid clearance by the reticuloendothelial system (RES) and/or degradation before reaching the target. In such instances, the complexes are swiftly cleared from circulation by the mononuclear phagocyte system (MPS), which results in the accumulation of complexes in off-target organs, such as the spleen and the liver, contributing to their distribution into healthy organs.49 Opsonization and recognition by immune cells, such as mononuclear phagocytes, pose a significant barrier to viral and non-viral delivery, as these professional phagocytic cells engulf the delivery particles before eliciting their effect.50,51
Afterward, the particles must cross the endothelium to exit the bloodstream and reach the target organ, tissue, or cell(s).
Once the delivery particles reach the target cells, they next encounter another significant barrier, namely the plasma membrane. Complexes often bind to the cell membrane through electrostatic interactions before entering the cell.52 Depending on their physicochemical features, complexes are internalized through passive (i.e., fusion) or active (receptor-mediated) transport. Endocytosis, defined as the process by which cells internalize substances through membrane-bound vesicles, is the primary mechanism responsible for the internalization of non-viral particles into cells. Regardless of the endocytic pathway (clathrin-mediated endocytosis (CME), caveolae-mediated endocytosis (CvME), or micropinocytosis),53 internalized polyplexes ultimately become trapped in endosomes. Therefore, another major hurdle to successful gene delivery is the release of the NA from the endosome before it undergoes degradation at the lysosomal level.54 This implies that the endosomal escape should be essential for efficient transfection. Different mechanisms have been suggested for polyplexes to escape the endosomal compartment. These rely on the proton sponge theory, and polyplex- or polymer-mediated membrane destabilization (for comprehensive reviews, refer to ref. 55 and 56).
According to the proton sponge theory, most polycations function as a proton buffer and their protonable groups cause osmotically-induced membrane burst.57 Upon endosome maturation, the membrane-bound V-ATPase proton pumps actively translocate H+ into the endosomal lumen. The polymers with high buffer capacity can thus bind H+, thereby counteracting endosome acidification. In an attempt to lower the pH, the proton pumps translocate even more H+ into the endosomal lumen, which goes along with the entry of Cl− ions to maintain the charge balance and leads to an increase in ionic concentration and an influx of water to maintain osmolarity. This process results in an increase in osmotic pressure that induces endosome swelling and eventually the release of the genetic cargo into the cytosol.58–62
In contrast to the proton sponge theory, which assumes that there is no interaction between polyplex and endosomal membrane, other studies support the theory of membrane permeability and nanohole formation.63,64 These theories explain the endosomal escape of CP-based vectors and differentiate between hole formation induced by an intact polyplex and through intercalations of free polymer with the endosomal membrane. In both cases, the endosomal compartment remains undamaged.65
In the first case, once entrapped within an endo-lysosomal compartment, the polyplexes will be protonated due to the activity of the membrane-bound ATPases. The polyplex with increasing charge density directly interacts with the phospholipids of the inner membrane leaflet. This interaction leads to membrane destabilization and a local loss of membrane integrity, thus forming nano-holes in it. The vector can escape the endo-lysosomal compartment through such holes.65,66 On the other hand, the free polymer-mediated escape theory is based on the unceasing exchange of the polymer chains between and to the polyplexes and the environment. Free polymer chains intercalate within the membrane of endo-lysosomal compartment, leading to membrane destabilization and a local loss of membrane integrity. The polyplex escapes the endo-lysosomal compartment because of the polymer-induced membrane destabilization,67 releasing the genetic cargo into the cytosol.
It is worth noting that while RNAs elicit their function in the cytosol, DNA molecules need to enter the nucleus. Hence, the intranuclear transport of DNA is another issue in the way of successful delivery.
Relying on the aforementioned considerations, an ideal NA vector shall: (i) be cationic to interact with NAs and give rise to positively charged complexes, which show enhanced NA protection and high affinity for the negatively charged cell surface; (ii) demonstrate buffer capacity to enable the proton-sponge effect; (iii) be biodegradable,68 meaning that they should be promptly eliminated from the body after the delivery of NAs. This behavior prevents significant accumulation in the body and minimizes potential organic damage, especially after repeated administrations.69
In the case of CPs, all these behaviors depend upon the structural and chemical features of the polymer, encompassing charge density, hydrophobicity/hydrophilicity, architecture, and molecular weight (Mw).70 In this regard, non-cationic building blocks have been utilized to endow CPs with improved DNA/RNA delivery behavior to tissues, cells, and subcellular compartments, while retaining stability in biological fluids.
Through the customization of the chemistry of these functional groups, we can enhance cell targeting behavior, uptake, endosomal escape, non-toxicity, and transfection efficiency. Furthermore, physicochemical properties such as loading capacity, complexation ability, size and morphology, biodegradability, pH sensitivity, and amphiphilicity can be tailored based on specific applications. This review comprehensively explores the role of non-cationic moieties in various biomedical applications, ranging from therapeutic interventions to gene editing. The rational incorporation of these building blocks for designing more effective CPs for NA delivery will be discussed hereafter.
With ongoing advancements in polymer chemistry, the invention and refinement of new polymerization techniques, along with the synthesis of novel functional monomers and post-functionalization methods, have enabled the constant development of polymeric nanomaterials for non-viral NA delivery. For a comprehensive overview of the state of the art of the synthetic methods used in these polymerization and post-functionalization techniques, we refer the reader to this more specialized literature.71–74
3. Cationic polymers as gene nanocarriers: an overview
CPs are commonly used in gene delivery applications36,75–77 due to their high packaging capacity, structural flexibility, and the ability for further functionalization.78,79 By adjusting their chemical composition,80,81 it is possible to control the release of cargo and ensure that CPs are degraded and eliminated from the body.82,83 CPs are made up of repeating units that contain (primary, secondary, and/or tertiary) amino groups as well as amidines, guanidines, and triazine moieties, which are responsible for their electrostatic interactions with the anionic NA phosphates to form polyplexes.21,84–86CPs employed for gene delivery applications are typically classified based on their origin as either synthetic or nature-derived polymers, and both categories are either commercially sourced or synthesized ad hoc.77,87
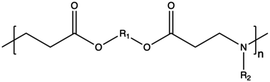
3.1. Synthetic CPs
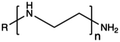
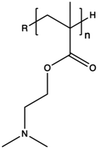
Synthetic polymers present precisely controlled chemical structures and appreciable ease of functionalization,88 as we can select a priori the building blocks that we consider the most suitable for their synthesis. As a result, they can ensure high batch-to-batch reproducibility and notable chemical versatility.89 The commercially sourced synthetic CPs most widely used in transfection include: (i) poly(ethyleneimine) (PEI),21,90–93 (ii) poly(amido amine) (PAMAM),94–97 (iii) poly(β-amino ester) (PβAEs),98–100 (iv) poly[2-(dimethylamino)ethyl methacrylate] (PDMAEMA),70,86,101,102 and (iv) poly(L-lysine) (PLL),103 which repeating units structures are reported in Table 1.Although PEI offers several advantages, such as low cost, ease of preparation and modification, and good transfection efficiency, its application is limited by drawbacks, including its high positive charge density, which can lead to cytotoxicity in high Mw (HMw) polymers.81,104,108,109 When comparing the cytotoxicity of equal concentrations of different PEIs (i.e., 800 Da bPEI, 25 kDa bPEI, and 20 kDa lPEI), bPEIs exhibited the highest toxicity. This heightened toxicity is attributed to the elevated density of methylene charge in the structure of bPEIs, leading to a greater destabilization of the cell membrane.110,111 On the other hand, experimental evidence shows that IPEI and bPEI with equivalent Mws exhibit comparable transfection efficiencies. Conversely, they demonstrate distinct NA complexation abilities, regardless of the Mw.77 Specifically, lPEI exclusively possesses secondary amines, whereas bPEI incorporates primary, secondary, and tertiary amines, depending on the polymerization degree.112 These structural differences account for the buffering capacity of bPEI across a broad pH range and, consequently, contribute to its “proton-sponge” behaviour. This effect, in turn, facilitates the escape of polyplexes from endo/lysosomes.
Under physiological pH conditions, all primary ε-amino groups of PLL are protonated, resulting in a structure lacking buffering capacity for endosomal escape.128 Typically, only PLLs with a Mw > 3 kDa can effectively condense DNA into stable complexes, underscoring the importance of the primary amine count. Despite the efficient condensation achieved by HMw PLL, these polymers often exhibit relatively high cytotoxicity.129
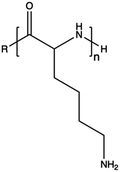
3.2. Natural polymers
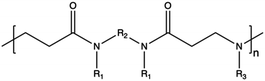
In contrast to synthetic CPs, nature-derived polymers have gained prominence because of their remarkable biocompatibility, bioactivity, and cell-mediated hydrolytic degradation.143,144 However, it remains challenging to accurately characterize and identify the chemical structure and composition of polymers obtained from natural resources. Therefore, such CPs are characterized by intrinsic batch-to-batch variability which complicates the development of new gene-delivery formulations.145,146 Furthermore, the high bioactivity of these natural polymers often triggers a robust immunogenic response.143 Natural CPs are typically sourced from two primary natural origins, namely proteins, biogenic polyamines, and polysaccharides.147 Chitosan (CS), a polysaccharide comprising randomly distributed β-(1-4)-linked D-glucosamine and N-acetyl-D-glucosamine, has been widely employed in gene delivery. This is due to its cationic nature, low toxicity, biodegradability, and minimal immunogenicity.148,149 However, CS also has some weaknesses, such as poor solubility under neutral pH conditions and lower NA complexation ability compared to other CPs.150,151 To address these issues, chemical modifications can be used to enhance the performance of CS, such as the formation of trimethyl CS.152 The reactive groups on the glucosamine units allow polysaccharides to undergo various chemical modifications,153 so that the resulting engineered derivatives can be customized to display specific biomedical functions.Spermine, a naturally occurring polyamine, assists in compacting cellular DNA within eukaryotic cells.154 However, on its own, it exhibits poor NA condensation, limited transfection efficiency due to its low Mw (∼200 Da), and poor endosomal escape capability, despite its good proton-buffering capacity.155,156 Studies have demonstrated that polymerizing spermine increasing its Mw can be advantageous for delivering siRNA.157 For instance, synthesis of spermine-based PβAEs revealed to be successful in obtaining polymers more biocompatible than 25 kDa bPEI and able to encapsulate and deliver siRNA more efficiently.158
4. Non-cationic building blocks for polycation modification
To date, various hybrid polycationic structures, integrating carefully selected functionalities, have been devised to enhance delivery efficiency, thus overcoming the biological barriers, biocompatibility, and interactions with target cells.159,160For example, modifying the surface properties of polycations through neutralization or hybridization with other biodegradable polymers or reducible groups can mitigate their toxicity,69,161,162 while enhancing in vivo bioavailability.163,164
On the other hand, enhanced biodegradability is a beneficial feature that facilitates the effective unpacking and release of loaded therapeutic genes in the cytosol.145,165 In this light, crosslinking or conjugating non-biodegradable polycations with stimuli-responsive functionalities can trigger carrier disruption. This, in turn, facilitates the controlled release of the cargo and subsequent elimination of the polymeric material from the bloodstream.58 Also, amine groups within the polymer chains can be utilized to anchor specific moieties and ligands to targeted particular organs and cell sites, limiting potential off-target delivery.166 The following sections highlight the most promising and updated strategies for incorporating non-cationic moieties, monomers, and functional groups into polycations, endowing them with specific behaviour. Table 2 showcases key examples of commonly employed functional unit structures.
| Property | Category | Monomers or functional units | Ref. |
|---|---|---|---|
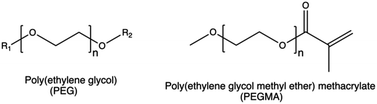
| Anti-opsonization | PEG chains |
|
174–176, 180, 222 and 223 |
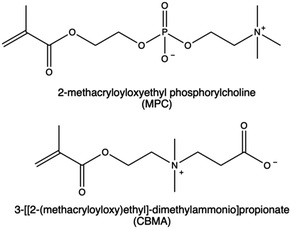
| Zwitterionic compounds |
|
182, 184–187, 189, 191 and 192 | |
| Poly(oxazoline)s |
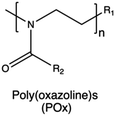
|
204, 205, 207, 210, 211, 214 and 219 | |
| Poly(phosphonate)s |
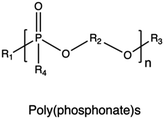
|
224–227 | |
| Advancing membrane-permeation | Alkyl chains |
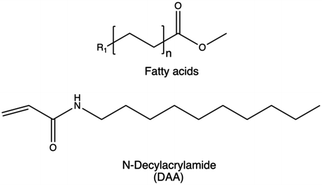
|
228–231 |
| Cholesterol derivatives |
|
232 and 233 | |
| Saponin derivatives |
|
234–238 | |
| Fluorine derivatives |
|
239–246 | |
| Penetrating peptides | Gly-Arg-Lys-Lys-Arg-Arg-Gln-Arg-Arg-Arg-Pro-Gln | 247–252 | |
| Transactivator of transcription (TAT) peptide | |||
| Arg-Gln-lle-Lys-lle-Trp-Phe-Gln-Asn-Arg-Arg-Met-Lys-Trp-Lys-Lys-Gly-Gly | |||
| Penetratin peptide | |||
| Gly-lle-Gly-Ala-Val-Leu-Lys-Val-Leu-Thr-Thr-Gly-Leu-Pro-Ala-Lue-lle-Ser-Trp-lle-Lys-Arg-Lys-Arg-Gln-Gln | |||
| Melittin peptide | |||
| Bioreducible | Disulphide bonds |
|
253–259 |
| Boronic acids |
|
260–262 | |
| Biodegradable | Synthetic |
|
263–267 |
| Polysaccharides |
|
268–271 | |
| Active targeting |
|
272–278 | |
| Branching |
|
279–284 |
4.1 Anti-opsonization functionality
PEG can induce an adverse immune reaction through the production of anti-PEG antibodies that, when present in high concentrations, lead to the rapid clearance of PEGylated complexes.115
Taking PEI as an example, PEG grafting onto the CP was shown to decrease the overall surface charge density, thereby affecting the cellular uptake, intracellular trafficking, and endosomal escape of PEI-based nanoparticles.176 Moreover, PEGylation can decrease the transfection efficiency of non-viral gene delivery systems by reducing the non-specific interactions between the cell and polyplexes.178 On the other hand, PEGylation increased the blood circulation time of PEI-based complexes,179 by reducing the interactions between polyplexes and the serum proteins.148,149,180,181
In this light, alternative surface modification strategies, including the use of different charge-shielding polymers, conditional removal of PEG, biomimetic surface modifications, and platelet membrane cloaking techniques, could potentially overcome the limitation of PEG.
Nonetheless, zwitterionic materials can also mitigate immune response and provide high stability to the nanovectors and prolong circulation time,193 thereby preventing the undesired long-term accumulation of nanocarriers in the RES.115,194 In literature, the development of CPs functionalized with zwitterionic moieties was primarily accomplished through the polymerization of 3-[[2-(methacryloyloxy)ethyl]-dimethylammonio]propionate (CBMA) and 2-methacryloyloxyethyl phosphorylcholine (MPC).195 Several studies have reported enhanced gene delivery efficacy and biocompatibility of PEI-based vectors following this functionalization.196,197
MPC derivatives containing a phosphorylcholine are highly water soluble198 and can undergo polymerization through various radical polymerization techniques.199 This allows for the production of cationic MPC copolymers with desired properties suitable for gene delivery purposes.200–203 In a work by Bhuchar et al., water-soluble CPs incorporating MPC and 2-aminoethyl methacrylamide hydrochloride (AEMA) with block and random architectures were synthesized by the reversible addition–fragmentation chain transfer (RAFT) method.203 As expected, the incorporation of MPC polymer segments in CPs significantly improved their biological performances. Polyplexes based on P(AEMA26-b-MPC27) were assessed in Hep G2 cells, and they were identified as optimal MPC copolymers due to their sustained low protein activation ability, combined with enhanced gene expression.203
In another study, a small library of zwitterion-modified PEIs was synthesized by grafting (in different ratios) 1,3-propanesultone or β-propiolactone onto hydrophobically modified (10 kDa and 25 kDa) bPEI (H-PEIs) via ring-opening addition reactions. The zwitterion moieties were introduced to H-PEIs via the reaction of amines producing N-sulfopropylated H-PEI (H-PEIs-S) and N-carboxyethylated H-PEI (H-PEIs-C), respectively. Evidence indicates that the zwitterionic modification conferred reduced cytotoxicity and mitigated nonspecific protein adsorption in the nanocarriers. However, no improvement was found in the overall siRNA to cancer cells (i.e., HeLa-Luc and A549-Luc cells). These results suggest that achieving an optimal balance between transfection efficacy and cytotoxicity may be dependent on the appropriate cationic to zwitterion components ratio.115
In addition, their water-solubility, anti-biofouling properties, cytocompatibility, and stealth effect227,285 explain the recent attention to developing novel synthetic methodologies and biomedical applications for PPEs.221 Indeed, numerous PPE-based drug delivery systems, PPE-containing nanomaterials for surface protein adsorption,286 and gene delivery nanocarriers225,287 have been examined. Notably, the interactions between hydrophilic PPEs and blood proteins have been widely explored.266,288 Schöttler et al. demonstrated that nanocarriers modified with either PEG or PPE exhibit reduced protein adsorption when incubated in human plasma compared to unmodified particles. Furthermore, mass spectrometric analysis of the protein corona formed on the particles’ surface post-plasma incubation revealed a similar protein profile for both PEGylated and ‘PPEylated’ particles.227 One of the first relevant study demonstrating the effectiveness of using PPE for gene delivery was proposed by Wang et al.289 The cationic copolymer was synthesized from poly(1,2-propylene phosphonate) by the Atherton–Todd reaction with spermidine side chain (PAA-SP). PPA-SP was able to efficiently condense plasmid DNA and formed complexes at N/P ratio ≥2. It offered notable protection to DNA against nuclease degradation and showed lower cytotoxicity than PLL and bPEI in cell culture. PPA-SP mediated efficient gene transfection in several cell lines, and the transfection protocol was optimized in HEK293 cells where PPA-SP/DNA polyplexes yield a luciferase expression level similar to that given by complexes made of PEI/DNA or based on TransFast™ commercial reagent.289 Previous studies demonstrated that amphiphilic cationic block copolymers of PEG, poly(ε-caprolactone) and poly(2-aminoethyl ethylene phosphate) (mPEG45-b-PCL30-b-PPEEA15) are capable of binding siRNA to form a micelle/siRNA complex (micelleplex).224 In another work, Mao et al. confirmed the in vivo efficiency of systemic administration of these micelleplexes for cancer therapy, by targeting the acid ceramidase (AC) oncogene, which plays a key role in cancer development.225,290 They investigated the knockdown in BT474 breast cancer cells and the inhibitory effect of micelleplexes administration on tumour growth. The micelleplexes obtained significantly reduce the expression of either a luciferase reporter gene or the endogenous acid ceramidase gene in cell culture, with acid ceramidase knockdown resulting in a significant increase in cell apoptosis. Importantly, this delivery system does not activate the innate immune response and is promising for systemic delivery of siRNAs for cancer therapy.225,287
4.2 Advancing membrane-permeation functionality
Linear alkyl chains of fatty acids including acetate, butanoate, hexanoate, myristate, and butyric anhydrides are the most commonly used hydrophobic groups as they increase colloidal stability and biocompatibility of the resulting nanovectors.228–230,297 However, the ultimate effect is significantly influenced by factors such as the degree of acetylation. Using bPEI as an example, an acetylation degree of 25% of primary amines was found to give the best overall transfection performaces.297 Similar results were observed when bPEI was functionalized with an acetylation degree of 57%, significantly improving cellular uptake performance compared to pristine bPEI 25 kDa.125 Conversely, a higher degree of acetylation was found to have a negative effect.126
Based on this observation, it can be concluded that a lower degree of functionalization is associated with better transfection performance. This may be attributed to the fact that acylation conceals the free amine groups of PEIs, thereby reducing their natural steric hindrance. The effect is significantly influenced by factors such as the degree of substitution, the position of the alkyl group, and the chain length. Similar observations rises from the copolymerization of HMw brushed poly(spermine acrylamides) (P(SpAA)) with N-decylacrylamide (DAA) monomers employed to increase the hydrophobicity of the resulting CPs.231 Different copolymers with varying ratios of cationic SpAA and hydrofobic DAA units were synthesized to investigate the impact of different hydrophobic fractions on siRNA delivery in presence of mucous. All the copolymers showed an improved cellular uptake in lung cells, and a ratio of 43/57 cationic to hydrophobic monomer subunits resulted in a better cellular uptake with respect to Lipofectamine and induced a ten-fold higher uptake than bPEI polyplexes under similar conditions.231
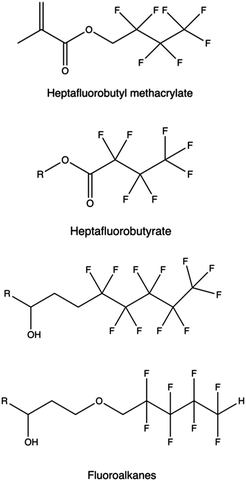
Alternatives fluorinated moieties commonly used to functionalize PAMAM, PPI, PEI, PLL, and other polymers are anhydrides, epoxides, ethyl esters, acyl chlorides, carboxyls, and acrylates.298
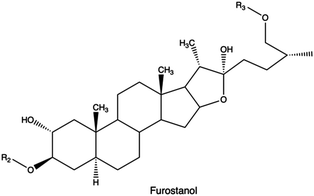
Saponins, which are glycosides found in plants and marine organisms comprised of a sugar residue attached to a steroidal or terpenoid aglycone,234 can function as a transfection enhancing agent. This is attributed to their membrane-permeabilizing function235 and ability to impart endosomal escape capabilities.236,237,313
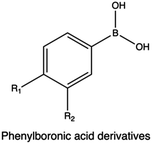
In a study Hasanzadeh et al., the bi-functionalization of LMw PEI with fluorine atoms and saponin residues resulted in a versatile and efficient delivery vehicle with reduced cytotoxicity. The authors utilized phenylboronic acid (PBA) as an ATP-responsive cross-linker for saponin-PEI conjugation. Complexes made of PEI-PBA-SAP-F and CRISPR/Cas9 system displayed good stability in serum and excellent gene editing ability in HEK 293T-GFP cells.238
4.3 Triggered disassembly
The incorporation of covalent bonds susceptible to degradation under physiological conditions presents an appealing approach to mitigate polymer toxicity, facilitate cargo release, and enhance transfection.314In the realm of biodegradable polymers, we distinguish between those that are inherently biodegradable (i.e., undergo spontaneous degradation) and stimuli-responsive ones, which experience triggered degradation in response to specific stimuli.
In the latter case, the modification involves introducing (bio)degradable (cross)links into the polycationic backbone or side chains, such as ester, imine, amide, carbamate, ketal, or disulfide bonds, enabling the creation of bio-reducible CPs.315 These nanomaterials undergo degradation into low Mw fragments through cleavage driven by chemical stimuli, such as pH variation, reactive oxygen species (ROS), adenosine triphosphate (ATP), and redox signals.316–319 As a result, the CPs effectively mitigate the issue of cytotoxicity while preserving the gene transfer potential of the pristine polymers.68,235–237,320
It is worthy of note that some stimuli, e.g. those driven by pH variations occurring when moving towards different intracellular environments (e.g., from cytosol to endosomes) or related to specific cellular activity, can be exploited to achieve the desired effect. This is evident when targeting the highly acidic tumor microenvironment.321 In this context, pH-responsive nanocarriers have been synthesized by incorporating ionizable pH-responsive functional groups, such as carboxylic acids, or acid-unstable covalent bonds, such as hydrazones, ester, orthoester, acetal, and cisacotinyl bonds.322–324 The resulting nanocarriers exhibit advantages in terms of cytotoxicity, biodegradability, cellular uptake, and NA release.147,325–327
A series of di-amine monomers containing stabilized ortho-ester groups were synthesized to give rise to an array of copolymers with unique pH-responsive properties.328 Moreover, Qi and co-workers obtained nanovectors based on pH-responsive branched polycations through the amino-epoxy ring-opening reaction of acid-sensitive monomers with impressive pH-responsive degradability in an acidic endosomal compartment (pH 5.0) and low-to-negligible cytotoxicity.329 This promoted the release of plasmid payload within the cytosol, with a subsequent increase in the transfection efficiency with respect to the non-pH-responsive counterparts.
ROS-cleavable delivery systems have been extensively reported in the literature. Several chemical groups, such as thioether, selenium, and PBA ester have been reported to be ROS responsive,330,331 undergoing physio-chemical and polarity variation in the presence of ROS. Thioacetals, instead, undergo direct cleavage under similar conditions. In a study by Zhang et al., the authors developed an array of fluorobenzene-substituted polycations with ROS-responsive thioacetal linkage via the diepoxide ring-opening polymerization and investigated the performances of the resulting polyplexes in terms of transfection efficiency and cytotoxicity in tumor (PC-3) cell lines. Interestingly, under higher ROS level conditions, polyplexes easily degrade while facilitating the intracellular release of DNA.332
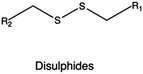
In the context of bio-reducible compounds, those containing disulfide bonds and boronic acid derivatives deserve special attention for the vast number of applications in gene delivery field.
Disulfide bonds have been efficiently incorporated in different CPs, including PEI, poly(amido ethylenimine) (PAEI), and poly(cystaminebisacrylamide-diaminohexane) (poly(CBA-DAH)).
In the preparation of bio-reducible PEI-based polymers, Gosselin et al. employed LMw PEI as the precursor, followed by cross-linking using two disulfide-containing cross-linkers, namely dithiobis-(succinimidylpropionate) (DSP) and dimethyl-3,3′-dithiobispropionimidate·2HCl (DTBP). These bio-reducible PEI-based polymers were used to prepare DNA-containing vectors that showed good transfection efficiencies in CHO cells owing to the stimulus-triggered release of the transgene via reductive cleavage of the disulfide bonds.253 In another study, Liu and collaborators synthesized a disulfide-containing cross-linked PEI (PEI-SS-CLs) using click chemistry. An azide-terminated PEI was initially synthesized from a LMw PEI (1.8 kDa), which was then converted into a HMw disulfide-containing cross-linked PEI through the introduction of a dialkyne cross-linker. The results showed that the cross-linked PEI exhibited both higher gene transfection efficiency and lower cytotoxicity if compared to the gold standard 25 kDa PEI.254
In an alternative strategy to obtain (bio)degradable CPs, Bauhuber and co-workers employed a smart chemistry approach to synthesize a library of PEG-SS-PEI diblock copolymers, where PEG moieties linked PEI through disulfide bonds, and compare them with non-degradable PEG-S-PEI (with a sulfide bond as the linker between PEG and PEI) copolymers.255 Interestingly, when tested at the same PEG contents, PEG-SS-PEI diblock copolymers were the most effective in transfection, highlighting the importance of the intracellular PEG cleavage in the transfection process. The same approach was employed to synthesize a novel bio-reducible targeted gene vector c(RGDyK)-poly(ethyleneglycol)-SS-PEI (RGD-PEG-SS-PEI), where a disulfide bond served as the linker for the PEG and PEI blocks. RGD-PEG-SS-PEI-based complexes carrying DNA showed higher transfection efficiency and reduced cytotoxicity both in vitro (in U87 cells) and in vivo (glioblastoma in mice) if compared with PEG-PEI complexes (no disulfide linkage).256
In the case of PAAs, Christensen et al. synthesized three types of disulfides containing PAEI from step polymerization of CBA and ethylenediamine (EDA), diethylenetriamine (DETA), and triethylenetetramine (TETA).257 These PAEI derivatives were capable of complexing DNA to form well-defined polyplexes with higher transfection efficiency than their pristine counterparts. In particular, the PAEIs resulting from CBA and TETA exhibited even higher transfection efficiency than 25 kDa bPEI even in the presence of serum. Chen and colleagues synthesized reducible hyperbranched PAEIs (RHBs) through Michael addition of N,N-dimethylamino dipropylenetriamine (DMDPTA), and two bisacrylamide monomers, CBA and HMBA, at specific molar feeding ratio to finely tune the density of disulfide linkages. When employed in transfection studies, the performances of these polymers as gene delivery vectors were dependent on the disulfide content, as this affected cytotoxicity, Mw of the degradation products, polyplex disassembly, and transfection efficiency.258 Similarly, Wu et al. synthesized PAAs with a controlled disulfide linkage content and studied their effect on PAEI cytotoxicity.259 Results indicated that the toxicity of nanovectors based on bio-reducible PAAs was decreased linearly with an increase in the disulfide content.
Interestingly, when evaluated in two cell lines, lower toxicity of bio-reducible PAEI-based complexes was observed in cells with higher levels of GSH. Jere et al. developed a bioresponsive reducible polyspermine (RPS) carrier composed of repeated spermine units with disulfide bridging obtained by Michael addition reaction between spermine and N,N′-cystaminebisacrylamide (CAM) in two stoichiometric ratios (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 4
1 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).343 RPS showed 1.8 times more resistance to pH change than bPEI 25 kDa thanks to the stronger buffering capacity, and strong DNA binding ability over a range of RPS/DNA ratios, indicating potential high transfection capabilities. Moreover, RPS demonstrated remarkable atoxicity even at concentrations 20-fold greater than those at which bPEI was cytotoxic. The study proved that RPS-pDNA binding is significantly reduced in the presence of a reducible environment, and RPS breakdown generates small spermine-like molecules easily metabolized and cleared by the cells.343 These results suggest that RPS might transfect cells more efficiently and safely than 25 kDa PEI.
1).343 RPS showed 1.8 times more resistance to pH change than bPEI 25 kDa thanks to the stronger buffering capacity, and strong DNA binding ability over a range of RPS/DNA ratios, indicating potential high transfection capabilities. Moreover, RPS demonstrated remarkable atoxicity even at concentrations 20-fold greater than those at which bPEI was cytotoxic. The study proved that RPS-pDNA binding is significantly reduced in the presence of a reducible environment, and RPS breakdown generates small spermine-like molecules easily metabolized and cleared by the cells.343 These results suggest that RPS might transfect cells more efficiently and safely than 25 kDa PEI.
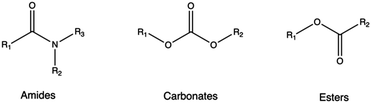
4.4 Hydrolytically labile functionality
Biodegradable synthetic polymers, such as polyesters, polyanhydrides, polycarbonates, polyamides, and polyurethanes,346 typically feature hydrolytically labile chemical bonds, such as esters, anhydrides, carbonates, amides, and urethanes, respectively.347As a rule of thumb, it is possible to confer biodegradability to non-biodegradable CPs by introducing specific moieties onto their structures. In these instances, non-cationic, naturally-derived biodegradable polymers, including dextran, cyclodextrin, and hyaluronic acid, can be employed for this purpose.153,348 Indeed, upon hydrolysis, these biodegradable polymers can be degraded into smaller components, which can be readily metabolized by the body.130
It is noteworthy that among the frequently used polycations listed in Table 1, PβAEs are notable for their degradation in both basic and acidic environments as well as physiological conditions. This degradation is facilitated by ester hydrolysis, resulting in the production of small bis(β-amino acid)s and diols, which are reported to possess relative biocompatibility without requiring further modification, thus circumventing issues related to the accumulation and toxicity.349 In contrast, all other CPs necessitate modifications to attain biodegradable characteristics.
The two major polymerization pathways used to incorporate non-cationic biodegradable structures to polycations are: (i) step polymerization, (ii) ring-opening polymerization (ROP), and (iii) a combination thereof.143,350–352
Modification of LMw PEIs with linear biodegradable polyesters was an effective strategy to improve the biocompatibility and degradability in physiological conditions.98 Indeed, the results of different studies showed that the complexes made of PEIs tethering polyester units were less cytotoxic and more effective in transfection if compared to the gold standard 25 kDa PEI.265,353 Similar outcomes were obtained with nanocarriers composed of 600 kDa PEI copolymerized with amino alcohol esters, diglycidyl adipate, diglycidyl succinate, and diglycidyl oxalate.341 Experimental evidence showed that, in comparison with 25 kDa PEI, these novel vectors facilitated DNA release, minimized cytotoxicity, and increased transfection efficiency.341
Poly(lactide-co-glycolide) (PLGA), polylactide (PLA), and poly(caprolactone) (PCL) are among the most widespread polyesters employed for polycation functionalization.98,263,350
Park and his team decorated PEI with PLGA units and achieved co-transfection in hMSCs cells by conjugating the SOX9 gene or core-binding factor alpha (cbfa-1) with small interfering RNA (siRNA) via ion interactions. In this way, they achieved good transfection efficiency and the desired nanocarrier biocompatibility features.265 In the study proposed by Xiong et al., the synthesis of a novel family of PEO-b-polyester copolymers grafted with short cationic moieties is reported.354 Poly(ethylene oxide)-b-poly(3-caprolactone-g-polyamine) (PEO-b-P(CL-g-polyamine)) block copolymers were prepared from PEO-b-poly(α-carboxyl-3-caprolactone) (PEO-b-PCCL), then, active ester method was used to attach pendant polyamine groups (as spermine (SP)) to the polyester section by amide bond formation. The developed amphiphilic copolymers were shown to be non-haemolytic, biodegradable, and less toxic than PEI against the chosen cancer cell line. They were also able to effectively bind siRNA, self-assemble into micelles and protect siRNA from degradation by nuclease in serum. PEO-b-P(CL-g-SP) micelles were very efficient in delivering siRNA into cytoplasm by endocytosis and facilitated endosome escape after cellular uptake.354
Biodegradable CPs were also developed using polyurethanes (PUs). Hung et al. synthesized an array of cationic PUs containing tertiary amines in the backbone and primary, secondary, or tertiary amines on the side chain. Such PUs were used to obtain biodegradable nanovectors with limited cytotoxicity and comparable transfection efficiency with respect to PEI and PDMAEMA.355 This experimental evidence supports the idea of using PUs combined with traditional polycarbonates (PCs) for in vivo gene delivery nanocarriers. In this regard, Liu et al. synthesized poly(ester-co-urethane) with 800 Da PEI on the side chain (PEU-g-PEI).356 In addition to its great biodegradability, PEU-g-PEI exhibited higher transfection efficiency in delivering DNA in COS-7 cells as compared with pristine PEI. Furthermore, Chiou and co-workers employed PU-PEI to deliver miRNA (miR145) in brain tumors357 and the PU-PEI-mediated miR145 delivery successfully inhibited the tumorigenesis of glioblastomas. They also employed the same PU-PEI carrier to treat lung cancer with a miR145-based therapy.358In vivo evidence showed promising results in xenograft tumors, including significantly reduced growth and metastasis, enhanced sensitivity of tumors to chemoradiotherapies, and prolonged survival of tumor-bearing mice. Furthermore, PU-PEI has been used for siRNA delivery to address metastasis in head and neck squamous cell carcinoma by targeting the two upregulated proteins, EZH2 and Oct4.359
Due to its high biocompatibility, low inherent cytotoxicity, and tuneable mechanical properties, PCs have emerged as an attractive alternative for gene delivery applications.360 He and co-workers initially synthesized poly(5-methyl-5-allyloxycarbonyl-trimethylene carbonate) (PMAC) using porcine pancreas lipase (IPPL).361 Subsequently, they converted the allyl functionality of PMAC to epoxide groups (PMAC-O), and further modified it with PEI to give rise to PMAC-g-PEI copolymers. When using PMAC-g-PEI to obtain gene delivery vectors, they showed enhanced transfection efficiency and lower cytotoxicity in 293T cells if compared to PEI-based polyplexes. Moreover, they additionally introduced 5,5-dimethyltrimethylene carbonate (DTC) into the backbone of PMAC-g-PEI, resulting in the preparation of P(MAC-co-DTC)-g-PEI which further affected the unpacking and endosomal escape of the loaded DNA.362
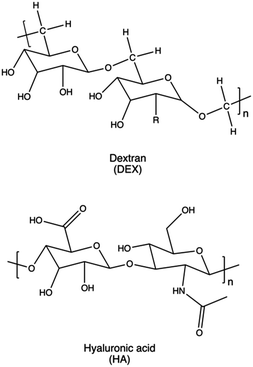
Dextran is a typical example of a biodegradable natural polysaccharide explored for gene delivery applications, owing to its remarkable water solubility and chemical flexibility.363 Dextran has been employed mainly to reduce the toxicity of PEI and other non-degradable polycations.160 Gong et al. reported efficient transfection and low cytotoxicity of a dextran-grafted-PEIs system, achieving miRNA transfer into osteosarcoma (OS) cells and OS-bearing nude mice.364 In a similar approach, Wang and colleagues modified the hydroxyl groups of dextran with a reduction-sensitive disulfide linker. This modification provided initiation sites for sequential PDMAEMA polymer chain growth through ATRP, aiming to reduce the overall cytotoxicity of the resulting delivery systems and further enhance their gene-transfection efficiency into various cell lines.365
4.5 Active-targeting functionality
Passive, active, or endogenous targeting may be used to facilitate the intracellular delivery of NAs to the appropriate organs, tissues, and cells.366 When active targeting is utilized, homing peptides, saccharides, antibodies, and proteins can be conjugated to polycations to bind to specific receptors and cellular compartments.367Immune cells, crucial in various diseases such as cancer and autoimmune disorders, are prime targets for NA delivery. Present throughout the body, they include macrophages, dendritic cells, and lymphocytes. Delivering genetic therapeutics to these cells offers potential for anti-inflammatory treatment, immune stimulation, and immunotherapies.367,368
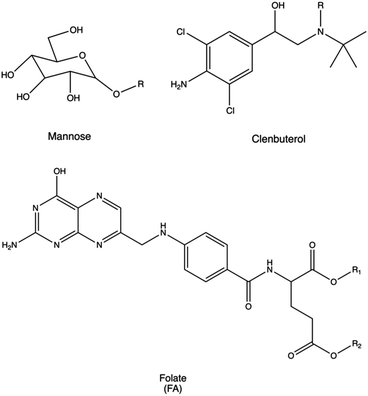
Macrophages can be targeted via their mannose receptor by conjugating mannose to PLL or polyaspartamide (pAsp(DET)).272,273 Using this approach, Man-PLL polyplexes displayed increased DNA transfection efficacy, and even higher transfection performances were observed when using Man-pAsp(DET) polyplexes to transfect murine bone marrow-derived macrophages. Similarly, dendritic cells were transfected with polyplexes made of mannose-PEI conjugates for targeted pDNA delivery, resulting in very high transfection efficacy.274
Primary human T cells were targeted via their CD3 receptor by conjugating anti-CD3 antibodies to PLL.369 In this way, pDNA polyplexes provided up to a 1000-fold increase in gene expression with respect to unmodified PLL.
Due to its intrinsic ability to accumulate nanoparticles, the liver is a crucial target for NA delivery. While a passive targeting approach may be used to accumulate the polymeric nanocarrier in this organ, specific targeting of hepatocytes might require additional synthetic efforts. This may be achieved by targeting the asialoglycoprotein receptor (ASGPR), which is highly expressed by hepatocytes.370 For instance, Plank et al. designed asialoorosomucoid-modified (ASOR) PLL-polyplexes to deliver pDNA to hepatocytes via ASGPR in vitro and in vivo.371
The lung is another crucial target organ which benefits from accessibility through both local and systemic routes. Overcoming the mucosal barrier and immune surveillance of alveolar sites is essential for the effective treatment of diseases such as cystic fibrosis, COPD, asthma, and pulmonary fibrosis, often necessitating receptor-targeting strategies.367 Lactoferrin receptor was typically targeted for the selective delivery of pDNA to bronchial epithelial cells.372 Accordingly, common approaches for lung targeting involve the conjugation of PEI with lactoferrin. Alternatively, other strategies utilize the conjugation with (i) the anti-platelet endothelial cell adhesion molecule (PECAM) antibody to achieve efficient systemic gene delivery to the lung with minimal toxicity,373 (ii) β2-adrenoceptor to deliver NAs into lung epithelial cells, (iii) clenbuterol to deliver pDNA and enhance gene expression in alveolar epithelial cells,275 (iv) prostaglandin I2 analogues (Iloprost and Treprostinil).374
Transferrin and lectins have also been investigated for lung targeting, as they were hypothesized to play a role in the specific nanocarrier uptake.375–378 In particular, targeted delivery of siRNA to pulmonary T cells via transferrin-PEI conjugated was investigated as a potential therapy for asthma.275,379,380
It is well known that treating the brain via systemic administration routes remains challenging due to the poor accessibility of the brain through the blood–brain barrier (BBB). Therefore, brain-targeted NA delivery may be achieved by decorating polyplexes with specific ligands that facilitate NA transfer via receptor-mediated transcytosis, which typically controls the permeation of nutrients, proteins, or lipids across the BBB.381
Transferrin and lactoferrin receptors, which regulate the transport of iron across the BBB, have been investigated for targeted delivery of PEG-PAMAM-382,383 and polymethylene polyphenylene isocyanate (PPI)-based384,385 nanocarriers to the central nervous system.386 Other strategies involve the conjugation with (i) retro-enantio peptide (re-TfR) to bind transferrin receptor and enhance transgene expression in TfR-rich K562 cells and Neuro2a cells,387 (ii) low-density lipoprotein receptor 1 (LRP1), angiopep and RVG29 peptides to facilitate overcoming BBB and the brain accumulation,381,386,388 (iii) peptide meningitis derived EPRNEEK for targeting laminin.389
Regarding tumor targeting, folate (folic acid, FA) is a widely employed ligand for anti-cancer agents because its target (folate receptor (FR)) is normally over-expressed in several cancer cells but rarely present on the surface of healthy cells.390 In particular, these RPs are elevated in malignant tissues of the ovary, uterus, endometrium, brain, kidney, head and neck, and skin.277 Liang et al. synthesized PEGylated PEI derivatives, where folate was conjugated on the distal end of the PEG-PEI complex (FA-PEG-PEI).278 They tested the effects of the polyplexes on three cell lines with different expression levels of FR (i.e., a FR-free hepatoblastoma HepG2 cell line, a low-level FR glioma C6 cell line, and a high-level FR 293T cell line). Compared with 25 kDa PEI, FA-PEG-PEI decreased the cytotoxicity and increased transgene expression in all the cell lines examined.278
Similarly, in a work proposed by Aranda et al., β-ciclodextrin conjugated with lPEI macromolecules were decorated with FA after polyplexes formation. The resulting nanocomplex showed to be efficient vectors for gene delivery in vivo in a mouse model, leading to relatively high transfection levels in the lung and, especially, in the liver.277
4.6 Branching functionality
The topological structures of polymers play a critical role in determining their performances as gene delivery vectors. Various non-linear polymers, including (hyper)branched, dendritic, multi-arm, and brush-shaped structures (Fig. 2), often display higher NA complexation ability than linear counterparts.142,391 Furthermore, non-linear polymer topologies have been found to affect the size and shape, charge density of the resulting polyplexes, and therefore their interactions with biological barriers and cellular compartments.142 | ||
| Fig. 2 Typical architectures of non-linear CPs employed for gene therapy (image created with Biorender.com). | ||
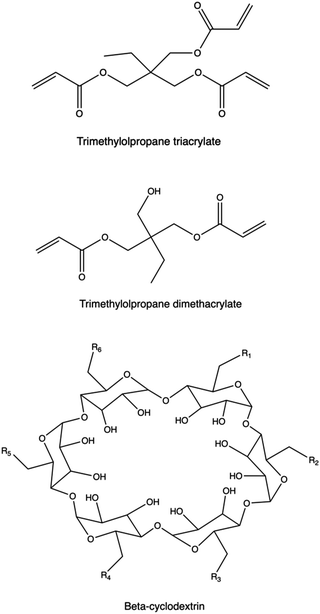
Introducing branching points into the polymer chain is essential to modify polymer topology. These units are either charged (e.g., tertiary amines) or neutral, and they can be incorporated during polymerization using multifunctional monomers or through post-modification methods, including crosslinking.158,391bPEI, PβAEs, PLL, PAMAM dendrimers present tertiary amines as branching points, and exhibit diverse amine groups with varying pKa values, enhancing NA binding and buffering capacity compared to linear polycations142,392–394 at the same Mw. However, higher charge density may increase cytotoxicity due to cell membrane disruption.395 Recent studies highlight the superior transfection efficiency and lower cytotoxicity of branched polymers like PβAEs, synthesized using non-ionic branching points from multifunctional acrylates via Michael addition of primary or secondary amines.122,349 PβAEs synthesized by adding non-ionic trimethylolpropane triacrylate as the multifunctional monomer were able to co-deliver plasmid DNA encoding Cas9 and short guide RNA (sgRNA), respectively, to enable gene knockout following a CRISPR-mediated cleavage at one genomic site as well as gene deletion following DNA cleavage.396 Recently, highly branched PAEs were investigated in SAR studies to highlight the effect of the polymer architecture on the gene transfection performances of the resulting polyplexes.
Xie et al. propose to employ citric acid (CA) as the backbone to link spermine (SPE) for developing a novel hyperbranched polyspermine (HPSPE).397 The resulting HPSPE was synthesized via a polymerization reaction between activated carboxyl groups of CA and amine groups of SPE, composed solely of endogenous compounds. HPSPE demonstrated efficient complexation of DNA into stable nanocomplexes with excellent biocompatibility profiles compared to 25 kDa bPEI, showing promising potential in addressing toxicity concerns associated with other synthetic materials. This is achieved while ensuring effective condensation of NAs due to the hyperbranched high Mw structure.397
A key structural factor, named branch unit distribution (BUD), was identified as playing an important role for transfection capability. Poly(amino esters) (PAEs) with a more uniform distribution of branch units, in fact, showed an enhanced transfection efficacy.398 Multi-arm, or “star” polymers are typically synthesized through controlled-living polymerization techniques, which allow to vary Mw and composition, depending on the specific application.399,400 The central core can be either a multifunctional LMw molecule or a macromolecule.142 PDMAEMA-based star polymers as well as other types of cationic polymers based on oligoethylene imine, poly(aminoethyl methacrylate) (PAEMA), PDMAEMA-block-poly(poly(ethylene glycol)ethyl ether methacrylate) (PPEGEEMA), have also been used as arms in the synthesis of star polymers via controlled polymerization for NA delivery.401,402
The use of cyclodextrins (CD) as central cores has been widely explored, via an effective conjugation of their multiple hydroxy groups to the polymeric arms,403–406 taking also advantage of their high biocompatibility, degradability, and possible encapsulation of other hydrophobic components in their inner compartment.407 Liu and colleagues synthesized a new CD derivative (CD-PLLD) consisting of a β-CD core and PLL dendron arms via click conjugation. Using CD-PLLD as the carrier, an anticancer drug docetaxel (DOC) and pMR3-specific siRNA can be co-loaded into a single carrier. With the co-loading of the hydrophobic drug and functional siRNA, CD-PLLD/DOC/pMR3 resulted in an enhanced apoptosis and a decreased invasive capacity of the treated HNE-1 cells than CD-PLLD/DOC (solely loading with DOC) and CD-PLLD/pMR3 (solely loading with pMR3 siRNA).408
Xiu et al. synthesized β-CD core-based cationic star polymers with PDMAEMA polymers as the multi-arm part via an ATRP technique. The resulting polymer can effectively condense pDNA and with this rational design gene-delivery system exhibited positive gene-transfection efficiency.409 The enhanced transfection performance revealed by star-shaped polycations obtained from a CD core was also confirmed in the work presented by Wang and collaborators. Linear poly(2-aminoethyl methacrylate) (l-PAEM) and three star-shaped PAEM polycations (s-PAEM) polymerized from β-CD at different Mw, were employed to condense pDNA. The transfection efficiency of the polyplexes was assessed in MCF-7 and COS-7 cells using pEGFP as the reporter gene.
Complexes formed by LMw-PAEMs exhibited the lowest cytotoxicity and, at a specific N/P ratio, the s-PAEMs showed significantly enhanced transfection efficiency than linear PAEMs of comparable Mw.410
Brush- or comb-like cationic polymers link several side chains into a single macromolecule. These macromolecular architectures may be exploited to improve the poor transfection efficiencies of oligocations, by grafting them to a common polymer backbone, thus achieving higher charge density.411,412 Graft polymers based on PDMAEMA, PEI, PEG-b-PEI, oligoamines, oligopeptides, and other structures have all been explored for NA delivery,142 with different type density and length of the polycationic grafts, as well as different controlled polymerization and post-polymerization strategies. Brush copolymers with pendant oligopeptide combs were obtained via free radical and RAFT polymerization of vinyl-terminated cationic oligopeptide, copolymerized with N-(2 hydroxypropyl) methacrylamide (HPMA).413
A post-polymerization of poly(glycidyl methacrylate) (PGMA) with tetraethylenepentamine (TEPA), pentaethylenehexamine (PEHA), and tris(2-aminoethyl) amine (TREN) was also conducted to generate a library of graft polymers for NA delivery.414 The ring-opening metathesis polymerization (ROMP) of cyclooctene-oligopeptide macromonomers resulted in comb-like structures exhibiting over two-fold higher pDNA transfection efficiency in COS-1 cells compared to non functionalized polyplexes. While their efficiency was lower than that of Lipofectamine, these structures demonstrated lower cytotoxicity.415 Tetralysine-containing comb polymers, modulated through copolymerization of a zwitterionic sulfobetaine-cyclooctene monomer, exhibited a weaker DNA binding affinity compared to the tetralysine-comb homopolymers. This resulted in a two-fold increase in delivery efficiency with SCOV3 cells.416
5. Conclusion and future perspectives
NAs have emerged as a new therapeutic category for preventing and treating various diseases. However, NA therapeutics must have safe, efficient, and biologically stable delivery systems to function effectively in vivo. Although ionizable lipid-based nanoparticles have shown promising clinical results in the two FDA-approved COVID-19 mRNA vaccines, in the FDA-approved liver-targeting siRNA formulation Onpattro®, and liver gene editing clinical trials,417 the restricted delivery to non-hepatic targets and tumors remains a big hurdle. Further optimization of non-viral synthetic delivery systems is thus urged. In this context, polymeric systems encompassing a number of tuneable moieties and functional groups offer a promising avenue for successful NA delivery.Polymeric vehicles for NA delivery have advanced significantly in the last decade, addressing challenges such as stability and intracellular trafficking. Stable polyplexes feature ionizable, polycationic moieties that enable electrostatic interactions with negatively charged NAs. Nonetheless, the presence of non-cationic components has been leveraged to create polycations with improved NA delivery, stability in biological fluids, and tailored physicochemical properties. Customizing macromolecular structures and innovative designs made it possible to refine toxicity-efficiency trade-offs and enhance precision targeting. Additionally, properties such as loading capacity and stimuli responsiveness can now be tailored to the specific application. Although viral vectors currently dominate the clinical gene therapy panorama, it is expected that synthetic vectors will increasingly prevail in clinical trials in the coming decades.418 Despite their promise, polyplex delivery systems are still in their early development stages. Indeed, according to the global clinical trial database (ref. ClinicalTrials.gov), there is only a limited number of human clinical trials involving polyplexes.419
This is primarily because of the layered complexity of these nanomaterials. Most polymeric delivery systems in use comprise quite complex formulations and cumbersome preparation to ensure good performance (e.g., stability, target specificity, and efficient delivery). Transitioning these systems from the laboratory scale to large-scale production for clinical use requires extensive optimization and standardization. Additionally, some delivery vehicles may trigger immune response and toxicity, and lead to off-target effects.
The slow progress in clinical translation may also be ascribed to inadequate screening workflows, which rely on the insufficient, inefficient, and non-robust methods used to assess the efficacy, safety, and other critical polyplex features. As a result, it is currently very challenging to identify the most effective and safe polyplex formulations, slowing down the overall progress of bringing them to the clinic and delaying benefits to patients. To overcome this issue, it is critically important to design and leverage high throughput in vitro platforms, which can significantly speed up and scale up preliminary screening for factors such as efficacy, toxicity, and stability.420–422 Similarly, high throughput in vivo platforms may enable the rapid assessment of how these formulations behave in complex biological systems, including their biodistribution, pharmacokinetics, and therapeutic effects. Moreover, coordinated pre-clinical studies may ensure that all necessary safety, efficacy, and regulatory requirements are met comprehensively and efficiently, providing robust and reliable data to pinpoint the best polyplex formulations for further development.
The majority of current clinical trials focus on mRNA-based therapeutics and vaccines.423,424 Advanced polyplex design may also lead to a shift in focus toward other applications, such as the delivery of ASOs and siRNA, both of which have demonstrated the ability to silence specific genes and offer potential treatments for a wide range of diseases. Just as an example, CRISPR/Cas9 is now the gold standard for therapeutic gene editing, but we still need to enhance its delivery efficiency to fully utilize its potential.
Besides, while some NA delivery systems have demonstrated some targeting effectiveness in vitro, in vivo targeting of tissues and organs remains a significant challenge. In this regard, a deeper understanding of biological processes and barriers governing cell-nanomaterial interactions, coupled with innovations in polymer chemistry, will continue to enable the development of advanced NA delivery systems. Modular synthesis accompanied by thorough characterization is a key area for improvement, and current advancements in polymer science hold promise for widespread clinical deployment in gene therapy. In this regard, the synthetic routes should ensure scalability and adherence to good manufacturing practices (GMP).
It is expected that personalized medicine will play a crucial role in the near future. Consequently, customizing polymeric vectors to match individual patient requirements, e.g., genotypic and phenotypic variations, will be essential. Polyplexes represent an exciting avenue for NA delivery and a valuable tool in the quest for effective treatments. With ongoing research, we can look forward to breakthroughs that will reshape medicine and improve patient outcomes.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.Conflicts of interest
There are no conflicts to declare.References
- T. K. Kim and J. H. Eberwine, Anal. Bioanal. Chem., 2010, 397, 3173–3178 CrossRef CAS PubMed.
- M. J. Lin, J. Svensson-Arvelund, G. S. Lubitz, A. Marabelle, I. Melero, B. D. Brown and J. D. Brody, Nat. Cancer, 2022, 3, 911–926 CrossRef CAS PubMed.
- N. Barda, N. Dagan, Y. Ben-Shlomo, E. Kepten, J. Waxman, R. Ohana, M. A. Hernán, M. Lipsitch, I. Kohane, D. Netzer, B. Y. Reis and R. D. Balicer, N. Engl. J. Med., 2021, 385, 1078–1090 CrossRef CAS PubMed.
- F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Pérez Marc, E. D. Moreira, C. Zerbini, R. Bailey, K. A. Swanson, S. Roychoudhury, K. Koury, P. Li, W. V. Kalina, D. Cooper, R. W. Frenck, L. L. Hammitt, Ö. Türeci, H. Nell, A. Schaefer, S. Ünal, D. B. Tresnan, S. Mather, P. R. Dormitzer, U. Şahin, K. U. Jansen and W. C. Gruber, N. Engl. J. Med., 2020, 383, 2603–2615 CrossRef CAS PubMed.
- E. E. Walsh, R. W. Frenck, A. R. Falsey, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, K. Neuzil, M. J. Mulligan, R. Bailey, K. A. Swanson, P. Li, K. Koury, W. Kalina, D. Cooper, C. Fontes-Garfias, P.-Y. Shi, Ö. Türeci, K. R. Tompkins, K. E. Lyke, V. Raabe, P. R. Dormitzer, K. U. Jansen, U. Şahin and W. C. Gruber, N. Engl. J. Med., 2020, 383, 2439–2450 CrossRef CAS PubMed.
- C. Wang, C. Pan, H. Yong, F. Wang, T. Bo, Y. Zhao, B. Ma, W. He and M. Li, J. Nanobiotechnol., 2023, 21 Search PubMed.
- J. Buck, P. Grossen, P. R. Cullis, J. Huwyler and D. Witzigmann, ACS Nano, 2019, 13 Search PubMed.
- P. J. Paddison, A. A. Caudy, E. Bernstein, G. J. Hannon and D. S. Conklin, Genes Dev., 2002, 16, 948–958 CrossRef CAS PubMed.
- C. B. Moore, E. H. Guthrie, M. T. H. Huang and D. J. Taxman, Methods Mol. Biol., 2010, 629, 141–158 Search PubMed.
- S. Bajan and G. Hutvagner, Cells, 2020, 9 Search PubMed.
- J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt, P. R. Cullis and R. van der Meel, Nat. Nanotechnol., 2021, 16 Search PubMed.
- D. M. Klinman, Nat. Rev. Immunol., 2004, 4, 249–258 CrossRef CAS PubMed.
- Z. Zhang, J. C. T. Kuo, S. Yao, C. Zhang, H. Khan and R. J. Lee, Pharmaceutics, 2022, 14 Search PubMed.
- T. Sugiyama, M. Gursel, F. Takeshita, C. Coban, J. Conover, T. Kaisho, S. Akira, D. M. Klinman and K. J. Ishii, J. Immunol., 2005, 174, 2273–2279 CrossRef CAS PubMed.
- T. Hui, W. Chou, S. Biswas and S. Lu, Methods Mol. Biol., 2004, 245 Search PubMed.
- A. J. Mellott, M. L. Forrest and M. S. Detamore, Ann. Biomed. Eng., 2013, 41, 446–468 CrossRef PubMed.
- D. J. Wells, Gene Ther., 2004, 11, 1363–1369 CrossRef CAS PubMed.
- J. T. Bulcha, Y. Wang, H. Ma, P. W. L. Tai and G. Gao, Signal Transduction Targeted Ther., 2021, 6 Search PubMed.
- M. A. Kay, J. C. Glorioso and L. Naldini, Nat. Med., 2001, 7, 33–40 CrossRef CAS PubMed.
- F. Ponti, M. Campolungo, C. Melchiori, N. Bono and G. Candiani, Chem. Phys. Lipids, 2021, 235, 105032 CrossRef CAS PubMed.
- N. Bono, F. Ponti, D. Mantovani and G. Candiani, Pharmaceutics, 2020, 12, 183 CrossRef CAS PubMed.
- J. L. Young and D. A. Dean, Adv. Genet., 2015, 89, 49–88 CAS.
- A. Delalande, C. Leduc, P. Midoux, M. Postema and C. Pichon, Ultrasound Med. Biol., 2015, 41, 1913–1926 CrossRef PubMed.
- L. Prosen, S. Prijic, B. Music, J. Lavrencak, M. Cemazar and G. Sersa, BioMed. Res. Int., 2013, 2013 DOI:10.1155/2013/209452.
- A. A. Davis, M. J. Farrar, N. Nishimura, M. M. Jin and C. B. Schaffer, Biophys. J., 2013, 105, 862–871 CrossRef CAS PubMed.
- K. Miller, A. L. Eggenberger, K. Lee, F. Liu, M. Kang, M. Drent, A. Ruba, T. Kirscht, K. Wang and S. Jiang, Sci. Rep., 2021, 11 DOI:10.1038/s41598-021-86549-9.
- Y. T. Chow, S. Chen, R. Wang, C. Liu, C. W. Kong, R. A. Li, S. H. Cheng and D. Sun, Sci. Rep., 2016, 6, 1–9 CrossRef PubMed.
- C. Li and R. J. Samulski, Nat. Rev. Genet., 2020, 21 Search PubMed.
- M. C. Milone and U. O'Doherty, Leukemia, 2018, 32, 1529–1541 CrossRef CAS PubMed.
- X. Li, Y. Le, Z. Zhang, X. Nian, B. Liu and X. Yang, Int. J. Mol. Sci., 2023, 24, 7736 CrossRef CAS PubMed.
- N. Singh and C. L. Heldt, Curr. Opin. Chem. Eng., 2022, 35, 100780 CrossRef.
- C. E. Thomas, A. Ehrhardt and M. A. Kay, Nat. Rev. Genet., 2003, 4, 346–358 CrossRef CAS PubMed.
- S. Ghosh, A. M. Brown, C. Jenkins and K. Campbell, Appl. Biosaf., 2020, 25, 7–18 CrossRef PubMed.
- I. Lostalé-Seijo and J. Montenegro, Nat. Rev. Chem., 2018, 2, 258–277 CrossRef.
- R. Ni, R. Feng and Y. Chau, Life, 2019, 9, 59 CrossRef CAS PubMed.
- M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259–302 CrossRef CAS PubMed.
- A. Aied, U. Greiser, A. Pandit and W. Wang, Drug Discovery Today, 2013, 18, 1090–1098 CrossRef CAS PubMed.
- W. T. Godbey and A. G. Mikos, J. Controlled Release, 2001, 72, 115–125 CrossRef CAS PubMed.
- C. J. Grimme, M. G. Hanson, L. G. Corcoran and T. M. Reineke, Biomacromolecules, 2022, 23, 3257–3271 CrossRef CAS PubMed.
- D. J. Gary, H. Lee, R. Sharma, J. S. Lee, Y. Kim, Z. Y. Cui, D. Jia, V. D. Bowman, P. R. Chipman, L. Wan, Y. Zou, G. Mao, K. Park, B. S. Herbert, S. F. Konieczny and Y. Y. Won, ACS Nano, 2011, 5, 3493–3505 CrossRef CAS PubMed.
- L. Zhou, M. Emenuga, S. Kumar, Z. Lamantia, M. Figueiredo and T. Emrick, Biomacromolecules, 2022, 23, 4029–4040 CrossRef CAS PubMed.
- D. Pack, A. Hoffman, S. Pun and P. Stayton, Nat. Rev. Drug Discovery, 2005, 4, 581–593 CrossRef CAS PubMed.
- M. P. Stewart, A. Sharei, X. Ding, G. Sahay, R. Langer and K. F. Jensen, Nature, 2016, 538, 183–192 CrossRef CAS PubMed.
- B. R. Olden, E. Cheng, Y. Cheng and S. H. Pun, Biomater. Sci., 2019, 7, 789–797 RSC.
- G. Lin, H. Zhang and L. Huang, Mol. Pharm., 2015, 12, 314–321 CrossRef CAS PubMed.
- S. M. Dizaj, S. Jafari and A. Y. Khosroushahi, Nanoscale Res. Lett., 2014, 9, 1–9 CrossRef CAS PubMed.
- O. M. Merkel and T. Kissel, Acc. Chem. Res., 2012, 45, 961–970 CrossRef CAS PubMed.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2020, 20, 101–124 CrossRef PubMed.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- F. Alexis, E. Pridgen, L. K. Molnar and O. C. Farokhzad, in Molecular Pharmaceutics, 2008, vol. 5, pp. 505–515 Search PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- Y. J. Ooi, Y. Wen, J. Zhu, X. Song and J. Li, Biomacromolecules, 2020, 21, 1136–1148 CrossRef CAS PubMed.
- A. El-Sayed and H. Harashima, Mol. Ther., 2013, 21, 1118–1130 CrossRef CAS PubMed.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS PubMed.
- T. Bus, A. Traeger and U. S. Schubert, J. Mater. Chem. B, 2018, 6, 6904–6918 RSC.
- B. Winkeljann, D. C. Keul and O. M. Merkel, J. Controlled Release, 2023, 353, 518–534 CrossRef CAS PubMed.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- I. M. S. Degors, C. Wang, Z. U. Rehman and I. S. Zuhorn, Acc. Chem. Res., 2019, 52, 1750–1760 CrossRef CAS PubMed.
- A. M. Funhoff, C. F. van Nostrum, G. A. Koning, N. M. E. Schuurmans-Nieuwenbroek, D. J. A. Crommelin and W. E. Hennink, Biomacromolecules, 2004, 5, 32–39 CrossRef CAS PubMed.
- L. M. P. Vermeulen, S. C. De Smedt, K. Remaut and K. Braeckmans, Eur. J. Pharm. Biopharm., 2018, 129, 184–190 CrossRef CAS PubMed.
- A. C. Rinkenauer, S. Schubert, A. Traeger and U. S. Schubert, J. Mater. Chem. B, 2015, 3, 7477–7493 RSC.
- Z. Zhou, X. Liu, D. Zhu, Y. Wang, Z. Zhang, X. Zhou, N. Qiu, X. Chen and Y. Shen, Adv. Drug Delivery Rev., 2017, 115, 115–154 CrossRef CAS PubMed.
- Y. J. Kwon, Acc. Chem. Res., 2012, 45, 1077–1088 CrossRef CAS PubMed.
- L. I. Selby, C. M. Cortez-Jugo, G. K. Such and A. P. R. Johnston, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9 Search PubMed.
- Z. U. Rehman, D. Hoekstra and I. S. Zuhorn, ACS Nano, 2013, 7, 3767–3777 CrossRef PubMed.
- C. H. Jones, C. K. Chen, A. Ravikrishnan, S. Rane and B. A. Pfeifer, Mol. Pharm., 2013, 10, 4082–4098 CrossRef CAS PubMed.
- S. Vaidyanathan, B. G. Orr and M. M. Banaszak Holl, Acc. Chem. Res., 2016, 49, 1486–1493 CrossRef CAS PubMed.
- Q. Y. Liu, R. Shi and L. Q. Zhang, Prog. Polym. Sci., 2012, 37, 715–765 CrossRef CAS.
- J. Luten, C. F. van Nostrum, S. C. De Smedt and W. E. Hennink, J. Controlled Release, 2008, 126, 97–110 CrossRef CAS PubMed.
- M. Foldvari, D. W. Chen, N. Nafissi, D. Calderon, L. Narsineni and A. Rafiee, J. Controlled Release, 2016, 240, 165–190 CrossRef CAS PubMed.
- F. J. Xu and W. T. Yang, Prog. Polym. Sci., 2011, 36, 1099–1131 CrossRef CAS.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. Van Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- N. Akeroyd and B. Klumperman, Eur. Polym. J., 2011, 47, 1207–1231 CrossRef CAS.
- C. K. Chen, P. K. Huang, W. C. Law, C. H. Chu, N. T. Chen and L. W. Lo, Int. J. Nanomed., 2020, 15, 2131–2150 CrossRef CAS PubMed.
- Y. Yue and C. Wu, Biomater. Sci., 2013, 1, 152–170 RSC.
- B. Shi, M. Zheng, W. Tao, R. Chung, D. Jin, D. Ghaffari and O. C. Farokhzad, Biomacromolecules, 2017, 18, 2231–2246 CrossRef CAS PubMed.
- C. Malloggi, D. Pezzoli, L. Magagnin, L. De Nardo, D. Mantovani, E. Tallarita and G. Candiani, Polym. Chem., 2015, 6, 6325–6339 RSC.
- J. W. Salameh, L. Zhou, S. M. Ward, C. F. Santa Chalarca, T. Emrick and M. L. Figueiredo, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2020, 12, 1–23 Search PubMed.
- H. Fang, Z. Guo, L. Lin, J. Chen, P. Sun, J. Wu, C. Xu, H. Tian and X. Chen, J. Am. Chem. Soc., 2018, 140, 11992–12000 CrossRef CAS PubMed.
- A. Gagliardi, E. Giuliano, E. Venkateswararao, M. Fresta, S. Bulotta, V. Awasthi and D. Cosco, Front. Pharmacol., 2021, 12 Search PubMed.
- J. Yang, K. M. Luly and J. J. Green, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2023, 15 Search PubMed.
- S. Vangala and G. Moku, Biomed. J. Sci. Tech. Res., 2021, 34, 26812–26815 Search PubMed.
- R. Rai, S. Alwani and I. Badea, Polymers, 2019, 11 Search PubMed.
- D. Pezzoli and G. Candiani, J. Nanopart. Res., 2013, 15 Search PubMed.
- D. Santo, R. A. Cordeiro, A. Sousa, A. Serra, J. F. J. Coelho and H. Faneca, Biomacromolecules, 2017, 18, 3331–3342 CrossRef CAS PubMed.
- A. Aied, U. Greiser, A. Pandit and W. Wang, Drug Discovery Today, 2013, 18, 1090–1098 CrossRef CAS PubMed.
- A. S. Piotrowski-Daspit, A. C. Kauffman, L. G. Bracaglia and W. M. Saltzman, Adv. Drug Delivery Rev., 2020, 156, 119–132 CrossRef CAS PubMed.
- J. Kim, H. Y. Jung and M. J. Park, Macromolecules, 2020, 53, 746–763 CrossRef CAS.
- M. I. Sabir, X. Xu and L. Li, J. Mater. Sci., 2009, 44, 5713–5724 CrossRef CAS.
- O. Boussif, F. Lezoualc'ht, M. A. Zanta, D. Mergnyt, D. Schermant, B. Demeneixt and J.-P. Behr, A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine, 1995, vol. 92 Search PubMed.
- I. Ullah, J. Zhao, B. Su, S. Rukh, J. Guo, X. Ren, S. Xia, W. Zhang and Y. Feng, J. Mater. Sci. Mater. Med., 2020, 31, 118 CrossRef CAS PubMed.
- D. Pezzoli, E. K. Tsekoura, K. C. Remant Bahadur, G. Candiani, D. Mantovani and H. Uludağ, Sci. China Mater., 2017, 60, 529–542 CrossRef CAS.
- D. Pezzoli, E. Giupponi, D. Mantovani and G. Candiani, Sci. Rep., 2017, 7 DOI:10.1038/srep44134.
- Y. Sun, H. Liu, H. Xing, L. Lang, L. Cheng, T. Yang, L. Yang and P. Ding, Polym. Int., 2019, 68, 447–455 CrossRef CAS.
- N. Bono, C. Pennetta, M. C. Bellucci, A. Sganappa, C. Malloggi, G. Tedeschi, G. Candiani and A. Volonterio, ACS Omega, 2019, 4, 6796–6807 CrossRef CAS.
- A. Ghilardi, D. Pezzoli, M. C. Bellucci, C. Malloggi, A. Negri, A. Sganappa, G. Tedeschi, G. Candiani and A. Volonterio, Bioconjugate Chem., 2013, 24, 1928–1936 CrossRef CAS PubMed.
- G. S. Yu, Y. M. Bae, H. Choi, B. Kong, I. S. Choi and J. S. Choi, Bioconjugate Chem., 2011, 22, 1046–1055 CrossRef CAS PubMed.
- T. J. Thomas, H. A. Tajmir-Riahi and C. K. S. Pillai, Molecules, 2019, 24 Search PubMed.
- T. R. Blake, W. C. Ho, C. R. Turlington, X. Zang, M. A. Huttner, P. A. Wender and R. M. Waymouth, Chem. Sci., 2020, 11, 2951–2966 RSC.
- J. Karlsson, K. R. Rhodes, J. J. Green and S. Y. Tzeng, Expert Opin. Drug Delivery, 2020, 17, 1395–1410 CrossRef CAS PubMed.
- H. Wang, Y. Jiang, H. Peng, Y. Chen, P. Zhu and Y. Huang, Adv. Drug Delivery Rev., 2015, 81, 142–160 CrossRef CAS PubMed.
- D. Santo, P. V. Mendonça, M. S. Lima, R. A. Cordeiro, L. Cabanas, A. Serra, J. F. J. Coelho and H. Faneca, Mol. Pharm., 2019, 16, 2129–2141 CrossRef CAS PubMed.
- M. A. Urello, L. Xiang, R. Colombo, A. Ma, A. Joseph, J. Boyd, N. Peterson, C. Gao, H. Wu and R. J. Christie, Biomacromolecules, 2020, 21, 3596–3607 CrossRef CAS PubMed.
- J. Lalani and A. Misra, in Challenges in Delivery of Therapeutic Genomics and Proteomics, Elsevier Inc., 2011, pp. 127–206 Search PubMed.
- X. Xu, B. Pejcic, C. Heath and C. Wood, J. Mater. Chem. A, 2018, 6, 21468–21474 RSC.
- Z. Zhang, X. Xiong, J. Wan, L. Xiao, L. Gan, Y. Feng, H. Xu and X. Yang, Biomaterials, 2012, 33, 7233–7240 CrossRef CAS PubMed.
- S. A. B. Riedl, P. Kaiser, A. Raup, C. V. Synatschke, V. Jérôme and R. Freitag, Processes, 2018, 6, 188 CrossRef CAS.
- Z. Zhang, W. Shen, J. Ling, Y. Yan, J. Hu and Y. Cheng, Nat. Commun., 2018, 9, 1377 CrossRef PubMed.
- T. Zhang, Y. Huang, X. Ma, N. Gong, X. Liu, L. Liu, X. Ye, B. Hu, C. Li, J. H. Tian, A. Magrini, J. Zhang, W. Guo, J. Xing, F. Bottini and X. J. Liang, Nano Lett., 2018, 18, 6301–6311 CrossRef CAS PubMed.
- A. Freund, Complexation of Cationic Polymers with Nucleic Acids for Gene Delivery, The University of Manchester, 2014 Search PubMed.
- X. Yong-Bing, C. Gui-Lin and G. Ming-Quan, Antioxidants, 2019, 8, 296 CrossRef PubMed.
- L. Melone, B. Rossi, N. Pastori, W. Panzeri, A. Mele and C. Punta, ChemPlusChem, 2015, 80, 1408–1415 CrossRef CAS PubMed.
- X. Zhang, K. Hong, Q. Sun, Y. Zhu and J. Du, Biomater. Sci., 2021, 9, 5275–5292 RSC.
- M. Rana, J. Biomater. Sci., Polym. Ed., 2021, 32, 1219–1249 CrossRef CAS PubMed.
- F. Liu, H. Su, M. Li, W. Xie, Y. Yan and Q. Shuai, Int. J. Mol. Sci., 2022, 23, 5014 CrossRef CAS PubMed.
- J. Zhang, M. Li, M. Wang, H. Xu, Z. Wang, Y. Li, B. Ding and J. Gao, J. Nanobiotechnol., 2021, 19, 135 CrossRef CAS PubMed.
- F. Alves Batista, S. B. Cunha Fontele, L. K. Beserra Santos, L. Alves Filgueiras, S. Quaresma Nascimento, J. M. de Castro e Sousa, J. C. Ramos Gonçalves and A. Nogueira Mendes, Mater. Today Commun., 2020, 22, 100762 CrossRef CAS.
- Y. Jia, D. Niu, Q. Li, H. Huang, X. Li, K. Li, L. Li, C. Zhang, H. Zheng, Z. Zhu, Y. Yao, X. Zhao, P. Li and G. Yang, Nanoscale, 2019, 11, 2008–2016 RSC.
- A. Schallon, C. V. Synatschke, V. Jérôme, A. H. E. Müller and R. Freitag, Biomacromolecules, 2012, 13, 3463–3474 CrossRef CAS PubMed.
- S. Agarwal, Y. Zhang, S. Maji and A. Greiner, This review will discuss recent efforts in design, synthesis, and performance of poly(2-dimethylaminoethyl methacrylate) (PDMAEMA) nanocomplexes with DNA, 2012, vol. 15 Search PubMed.
- M. Zeng, Q. Xu, D. Zhou, A. Sigen, F. Alshehri, I. Lara-Sáez, Y. Zheng, M. Li and W. Wang, Adv. Drug Delivery Rev., 2021, 176 Search PubMed.
- R. A. Cordeiro, A. Serra, J. F. J. Coelho and H. Faneca, J. Controlled Release, 2019, 310, 155–187 CrossRef CAS PubMed.
- D. Shenoy, S. Little, R. Langer and M. Amiji, Mol. Pharm., 2005, 2, 357–366 CrossRef CAS PubMed.
- G. T. Zugates, N. C. Tedford, A. Zumbuehl, S. Jhunjhunwala, C. S. Kang, L. G. Griffith, D. A. Lauffenburger, R. Langer and D. G. Anderson, Bioconjugate Chem., 2007, 18, 1887–1896 CrossRef CAS PubMed.
- S. Perni and P. Prokopovich, Nanomedicine, 2017, 13, 539–548 CrossRef CAS PubMed.
- J. C. Sunshine, D. Y. Peng and J. J. Green, Mol. Pharm., 2012, 9, 3375–3383 CrossRef CAS PubMed.
- C.-H. Ahn, S. Y. Chae, H. Bae and S. W. Kim, Biodegradable poly(ethylenimine) for plasmid DNA delivery, 2002, vol. 80 Search PubMed.
- A. Akinc and R. Langer, Biotechnol. Bioeng., 2002, 78, 503–508 CrossRef CAS PubMed.
- H. Lee, J. H. Jeong and T. G. Park, J. Controlled Release, 2002, 79, 283–291 CrossRef CAS PubMed.
- Y. Li, D. Maciel, J. Rodrigues, X. Shi and H. Tomás, Chem. Rev., 2015, 115, 8564–8608 CrossRef CAS PubMed.
- A. Frère, A. Baroni, E. Hendrick, A. S. Delvigne, F. Orange, O. Peulen, G. R. Dakwar, J. Diricq, P. Dubois, B. Evrard, K. Remaut, K. Braeckmans, S. C. De Smedt, J. Laloy, J. M. Dogné, G. Feller, L. Mespouille, D. Mottet and G. Piel, ACS Appl. Mater. Interfaces, 2017, 9, 2181–2195 CrossRef PubMed.
- D. Hwang, J. D. Ramsey, N. Makita, C. Sachse, R. Jordan, M. Sokolsky-Papkov and A. V. Kabanov, J. Controlled Release, 2019, 307, 261–271 CrossRef CAS PubMed.
- J. Bai, J. Wang, Y. Feng, Y. Yao and X. Zhao, Colloids Surf., A, 2022, 639 DOI:10.1016/j.colsurfa.2022.128353.
- Y. Zhang, J. Shi, B. Ma, Y. N. Zhou, H. Yong, J. Li, X. Kong and D. Zhou, Prog. Polym. Sci., 2023, 146 Search PubMed.
- M. Hashemi, S. M. Tabatabai, H. Parhiz, S. Milanizadeh, S. A. Farzad, K. Abnous and M. Ramezani, Mater. Sci. Eng., C, 2016, 61, 791–800 CrossRef CAS PubMed.
- Y. Tang, Y. Liu, Y. Xie, J. Chen and Y. Dou, Asian J. Pharm. Sci., 2020, 15, 121–128 CrossRef PubMed.
- J. Karlsson, S. Y. Tzeng, S. Hemmati, K. M. Luly, O. Choi, Y. Rui, D. R. Wilson, K. L. Kozielski, A. Quiñones-Hinojosa and J. J. Green, Adv. Funct. Mater., 2021, 31 DOI:10.1002/adfm.202009768.
- B. A. Paray, A. Ahmad, J. M. Khan, F. Taufiq, A. Pathan, A. Malik and M. Z. Ahmed, Drug Discovery Today, 2021, 26, 1053–1059 CrossRef CAS PubMed.
- M. Meyer, A. Zintchenko, M. Ogris and E. Wagner, J. Gene Med., 2007, 9, 797–805 CrossRef CAS PubMed.
- R. Kandil, Y. Xie, R. Heermann, L. Isert, K. Jung, A. Mehta and O. M. Merkel, Adv. Ther., 2019, 2 DOI:10.1002/adtp.201900047.
- D. P. Feldmann, Y. Cheng, R. Kandil, Y. Xie, M. Mohammadi, H. Harz, A. Sharma, D. J. Peeler, A. Moszczynska, H. Leonhardt, S. H. Pun and O. M. Merkel, J. Controlled Release, 2018, 276, 50–58 CrossRef CAS PubMed.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. Van Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527–11652 CrossRef CAS PubMed.
- L. S. Nair and C. T. Laurencin, Prog. Polym. Sci., 2007, 32, 762–798 CrossRef CAS.
- C. K. Chen, W. J. Lin, Y. Hsia and L. W. Lo, Macromol. Biosci., 2017, 17 DOI:10.1002/mabi.201600191.
- S. Doppalapudi, A. Jain, W. Khan and A. J. Domb, Polym. Adv. Technol., 2014, 25, 427–435 CrossRef CAS.
- M. Vert, Int. J. Artif. Organs, 2011, 34, 76–83 CrossRef CAS PubMed.
- Y. Wang, S. Zhang and D. S. W. Benoit, J. Controlled Release, 2018, 287, 58–66 CrossRef CAS PubMed.
- I. Pilipenko, V. Korzhikov-Vlakh, V. Sharoyko, N. Zhang, M. Schäfer-Korting, E. Rühl, C. Zoschke and T. Tennikova, Pharmaceutics, 2019, 11 DOI:10.3390/pharmaceutics11070317.
- B. Cheng, H. H. Ahn, H. Nam, Z. Jiang, F. J. Gao, I. Minn and M. G. Pomper, Pharmaceutics, 2022, 4 DOI:10.3390/pharmaceutics14020373.
- Y. Ping, C. De Liu, G. P. Tang, J. S. Li, J. Li, W. T. Yang and F. J. Xu, Adv. Funct. Mater., 2010, 20, 3106–3116 CrossRef CAS.
- C. K. Chen and S. C. Huang, Mol. Pharm., 2016, 13, 4152–4167 CrossRef CAS PubMed.
- T. Kean, S. Roth and M. Thanou, J. Controlled Release, 2005, 103, 643–653 CrossRef CAS PubMed.
- W. W. Hu, C. C. Yeh and C. W. Tsai, J. Mater. Chem. B, 2018, 6, 5781–5794 RSC.
- A. Estévez-Torres and D. Baigl, Soft Matter, 2011, 7, 6746–6756 RSC.
- M. Elsayed, V. Corrand, V. Kolhatkar, Y. Xie, N. H. Kim, R. Kolhatkar and O. M. Merkel, Biomacromolecules, 2014, 15, 1299–1310 CrossRef CAS PubMed.
- J. G. Hardy, M. A. Kostiainen, D. K. Smith, N. P. Gabrielson and D. W. Pack, Bioconjugate Chem., 2006, 17, 172–178 CrossRef CAS PubMed.
- A. R. Lote, V. R. Kolhatkar, T. Insley, P. Král and R. Kolhatkar, ACS Macro Lett., 2014, 3, 829–833 CrossRef CAS PubMed.
- Y. Jin, F. Adams, L. Isert, D. Baldassi and O. M. Merkel, Mol. Pharm., 2023, 20, 4505–4516 CrossRef CAS PubMed.
- A. Zakeri, M. A. J. Kouhbanani, N. Beheshtkhoo, V. Beigi, S. M. Mousavi, S. A. R. Hashemi, A. K. Zade, A. M. Amani, A. Savardashtaki, E. Mirzaei, S. Jahandideh and A. Movahedpour, Nano Rev. Exp., 2018, 9, 1488497 CrossRef PubMed.
- C. K. Chen, P. K. Huang, W. C. Law, C. H. Chu, N. T. Chen and L. W. Lo, Int. J. Nanomed., 2020, 15, 2131–2150 CrossRef CAS PubMed.
- M. A. Islam, T. E. Park, B. Singh, S. Maharjan, J. Firdous, M. H. Cho, S. K. Kang, C. H. Yun, Y. J. Choi and C. S. Cho, J. Controlled Release, 2014, 193, 74–89 CrossRef CAS PubMed.
- Y. Xiang, N. N. L. Oo, J. P. Lee, Z. Li and X. J. Loh, Drug Discovery Today, 2017, 22, 1318–1335 CrossRef CAS PubMed.
- K. Kim, W. C. W. Chen, Y. Heo and Y. Wang, Prog. Polym. Sci., 2016, 60, 18–50 CrossRef CAS.
- L. Xie, Y. Tan, Z. Wang, H. Liu, N. Zhang, C. Zou, X. Liu, G. Liu, J. Lu and H. Zheng, ACS Appl. Mater. Interfaces, 2016, 8, 29261–29269 CrossRef CAS PubMed.
- R. B. Arote, H. L. Jiang, Y. K. Kim, M. H. Cho, Y. J. Choi and C. S. Cho, Expert Opin. Drug Delivery, 2011, 8, 1237–1246 CrossRef CAS PubMed.
- R. Masoumzadeh and A. Zare, Adv. Appl. NanoBio-Technol., 2021, 2, 13–16 CAS.
- M. Li, S. Jiang, J. Simon, D. Paßlick, M. L. Frey, M. Wagner, V. Mailander, D. Crespy and K. Landfester, Nano Lett., 2021, 21, 1591–1598 CrossRef CAS PubMed.
- M. Ahmed, N. Bhuchar, K. Ishihara and R. Narain, Bioconjugate Chem., 2011, 22, 1228–1238 CrossRef CAS PubMed.
- B. Le Bon, N. Van Craynest, O. Boussif and P. Vierling, Bioconjugate Chem., 2002, 13, 1292–1301 CrossRef CAS PubMed.
- T. Blessing, M. Kursa, R. Holzhauser, R. Kircheis and E. Wagner, Bioconjugate Chem., 2001, 12, 529–537 CrossRef CAS PubMed.
- H. Otsuka, Y. Nagasaki and K. Kataoka, Adv. Drug Delivery Rev., 2003, 55, 403–419 CrossRef CAS PubMed.
- T. Gillich, C. Acikgöz, L. Isa, A. D. Schlüter, N. D. Spencer and M. Textor, ACS Nano, 2013, 7, 316–329 CrossRef CAS PubMed.
- R. K. Kainthan and D. E. Brooks, Biomaterials, 2007, 28, 4779–4787 CrossRef CAS PubMed.
- K. Shiraishi and M. Yokoyama, Sci. Technol. Adv. Mater., 2019, 20, 324–336 CrossRef CAS PubMed.
- G. Osman, J. Rodriguez, S. Y. Chan, J. Chisholm, G. Duncan, N. Kim, A. L. Tatler, K. M. Shakesheff, J. Hanes, J. S. Suk and J. E. Dixon, J. Controlled Release, 2018, 285, 35–45 CrossRef CAS PubMed.
- Y. Fang, J. Xue, S. Gao, A. Lu, D. Yang, H. Jiang, Y. He and K. Shi, Drug Delivery, 2017, 24, 22–32 CrossRef CAS PubMed.
- M. Shen, L. Martinson, M. S. Wagner, D. G. Castner, B. D. Ratner and T. A. Horbett, J. Biomater. Sci., Polym. Ed., 2002, 13, 367–390 CrossRef CAS PubMed.
- G. Zhao, L. Long, L. Zhang, M. Peng, T. Cui, X. Wen, X. Zhou, L. Sun and L. Che, Sci. Rep., 2017, 7, 3383 CrossRef PubMed.
- A. Li, J. Qiu, B. Zhou, B. Xu, Z. Xiong, X. Hao, X. Shi and X. Cao, Arabian J. Chem., 2020, 13, 2558–2567 CrossRef CAS.
- J. Wang, H. Wang, P. Zhao, Z. Chen and Q. Lin, Colloids Interface Sci. Commun., 2022, 5, 100647 CrossRef.
- X. Y. Wang, T. Ishida and H. Kiwada, J. Controlled Release, 2007, 119, 234–244 Search PubMed.
- C. Leng, H. C. Hung, S. Sun, D. Wang, Y. Li, S. Jiang and Z. Chen, ACS Appl. Mater. Interfaces, 2015, 7, 16881–16888 CrossRef CAS PubMed.
- Q. Shao and S. Jiang, Adv. Mater., 2015, 27, 15–26 CrossRef CAS PubMed.
- M. He, K. Gao, L. Zhou, Z. Jiao, M. Wu, J. Cao, X. You, Z. Cai, Y. Su and Z. Jiang, Acta Biomater., 2016, 40, 142–152 CrossRef CAS PubMed.
- H. Zou, Z. Wang and M. Feng, J. Controlled Release, 2015, 214, 121–133 CrossRef CAS PubMed.
- J. B. Schlenoff, Langmuir, 2014, 30, 9625–9636 CrossRef CAS PubMed.
- C. Leng, S. Sun, K. Zhang, S. Jiang and Z. Chen, Acta Biomater., 2016, 40, 6–15 CrossRef CAS.
- L. Y. Zhou, Y. H. Zhu, X. Y. Wang, C. Shen, X. W. Wei, T. Xu and Z. Y. He, Comput. Struct. Biotechnol. J., 2020, 18, 1980–1999 CrossRef CAS.
- Y. He, J. Hower, S. Chen, M. T. Bernards, Y. Chang and S. Jiang, Langmuir, 2008, 24, 10358–10364 CrossRef CAS PubMed.
- D. Faulón Marruecos, M. Kastantin, D. K. Schwartz and J. L. Kaar, Biomacromolecules, 2016, 17, 1017–1025 CrossRef PubMed.
- B. Pelaz, P. Del Pino, P. Maffre, R. Hartmann, M. Gallego, S. Rivera-Fernández, J. M. De La Fuente, G. U. Nienhaus and W. J. Parak, ACS Nano, 2015, 9, 6996–7008 CrossRef CAS.
- N. Gal, M. Schroffenegger and E. Reimhult, J. Phys. Chem. B, 2018, 122, 5820–5834 CrossRef CAS.
- M. Harijan and M. Singh, J. Mol. Recognit., 2022, 35 Search PubMed.
- Z. Xiong, M. Shen and X. Shi, Bioconjugate Chem., 2019, 30, 2519–2527 CrossRef CAS PubMed.
- J.-L. Zhao, A. Lerner, Z. Shu, X.-J. Gao and C.-S. Zee, Radiol. Infect. Dis., 2015, 1, 94–102 CrossRef.
- K. M. Xiu, N. N. Zhao, W. T. Yang and F. J. Xu, Acta Biomater., 2013, 9, 7439–7448 CrossRef CAS PubMed.
- J. Sun, F. Zeng, H. Jian and S. Wu, Biomacromolecules, 2013, 14, 728–736 CrossRef CAS PubMed.
- K. Ishihara, T. Ueda and N. Nakabayashi, Preparation of Phospholipid Polymers and Their Properties as Polymer Hydrogel Membranes, 1990, vol. 22 Search PubMed.
- T. Ueda, H. Oshida, K. Kurita, K. Ishihara and N. Nakabayashi, Preparation of 2-Methacryloyloxyethyl Phosphorylcholine Copolymers with Alkyl Methacrylates and Their Blood Compatibility, 1992, vol. 24 Search PubMed.
- K. Ishihara, S. Hachiya, Y. Inoue, K. Fukazawa and T. Konno, Polymers, 2020, 12 DOI:10.3390/polym12081762.
- Y. T. A. Chim, J. K. W. Lam, Y. Ma, S. P. Armes, A. L. Lewis, C. J. Roberts, S. Stolnik, S. J. B. Tendler and M. C. Davies, Langmuir, 2005, 21, 3591–3598 CrossRef CAS.
- J. K. W. Lam, Y. Ma, S. P. Armes, A. L. Lewis, T. Baldwin and S. Stolnik, J. Controlled Release, 2004, 100, 293–312 CrossRef CAS.
- N. Bhuchar, Z. Deng, K. Ishihara and R. Narain, Polym. Chem., 2011, 2, 632–639 RSC.
- N. Adams and U. S. Schubert, Adv. Drug Delivery Rev., 2007, 59, 1504–1520 CrossRef CAS.
- R. Hoogenboom, Angew. Chem., Int. Ed., 2009, 48, 7978–7994 CrossRef CAS PubMed.
- H. Schlaad, C. Diehl, A. Gress, M. Meyer, A. Levent Demirel, Y. Nur and A. Bertin, Macromol. Rapid Commun., 2010, 31, 511–525 CrossRef CAS PubMed.
- R. Luxenhofer, Y. Han, A. Schulz, J. Tong, Z. He, A. V. Kabanov and R. Jordan, Macromol. Rapid Commun., 2012, 33, 1613–1631 CrossRef CAS PubMed.
- H. M. L. Lambermont-Thijs, F. S. Van Der Woerdt, A. Baumgaertel, L. Bonami, F. E. Du Prez, U. S. Schubert and R. Hoogenboom, Macromolecules, 2010, 43, 927–933 CrossRef CAS.
- M. Hartlieb, K. Kempe and U. S. Schubert, J. Mater. Chem. B, 2015, 3, 526–538 RSC.
- E. Haladjova, S. Rangelov and C. Tsvetanov, Polymers, 2020, 12, 1–18 CrossRef PubMed.
- E. Haladjova, M. Smolíček, I. Ugrinova, D. Momekova, P. Shestakova, Z. Kroneková, J. Kronek and S. Rangelov, J. Appl. Polym. Sci., 2020, 37 DOI:10.1002/app.49400.
- H. Bludau, A. E. Czapar, A. S. Pitek, S. Shukla, R. Jordan and N. F. Steinmetz, Eur. Polym. J., 2017, 88, 679–688 CrossRef CAS PubMed.
- M. Bauer, C. Lautenschlaeger, K. Kempe, L. Tauhardt, U. S. Schubert and D. Fischer, Macromol. Biosci., 2012, 12, 986–998 CrossRef CAS PubMed.
- R. Luxenhofer, G. Sahay, A. Schulz, D. Alakhova, T. K. Bronich, R. Jordan and A. V. Kabanov, in Journal of Controlled Release, 2011, vol. 153, pp. 73–82 Search PubMed.
- A. Mero, G. Pasut, L. D. Via, M. W. M. Fijten, U. S. Schubert, R. Hoogenboom and F. M. Veronese, J. Controlled Release, 2008, 125, 87–95 CrossRef CAS PubMed.
- M. Mees, E. Haladjova, D. Momekova, G. Momekov, P. S. Shestakova, C. B. Tsvetanov, R. Hoogenboom and S. Rangelov, Biomacromolecules, 2016, 17, 3580–3590 CrossRef CAS PubMed.
- T. Ivanova, E. Haladjova, M. Mees, D. Momekova, S. Rangelov, G. Momekov and R. Hoogenboom, Pharmacia, 2016, 63, 3–8 Search PubMed.
- N. Toncheva-Moncheva, E. Veleva-Kostadinova, C. Tsvetanov, D. Momekova and S. Rangelov, Polymer, 2017, 111, 156–167 CrossRef CAS.
- A. B. Cook, R. Peltier, J. Zhang, P. Gurnani, J. Tanaka, J. A. Burns, R. Dallmann, M. Hartlieb and S. Perrier, Polym. Chem., 2019, 10, 1202–1212 RSC.
- C. Pelosi, M. R. Tinè and F. R. Wurm, Eur. Polym. J., 2020, 141 Search PubMed.
- T. Steinbach and F. R. Wurm, Angew. Chem., 2015, 127, 6196–6207 CrossRef.
- K. S. Cho, S. Kim, H. B. Chun, J. H. Cheon, M. H. Cho, A. Y. Lee and R. B. Arote, PLoS One, 2022, 17 DOI:10.1371/journal.pone.0266181.
- T. P. Loginova, I. A. Khotina, Y. A. Kabachii, S. Y. Kochev, V. M. Abramov, V. S. Khlebnikov, N. L. Kulikova and Y. O. Mezhuev, Polymers, 2023, 15, 3036 CrossRef CAS PubMed.
- T. M. Sun, J. Z. Du, L. F. Yan, H. Q. Mao and J. Wang, Biomaterials, 2008, 29, 4348–4355 CrossRef CAS PubMed.
- C. Q. Mao, J. Z. Du, T. M. Sun, Y. D. Yao, P. Z. Zhang, E. W. Song and J. Wang, Biomaterials, 2011, 32, 3124–3133 CrossRef CAS.
- D. A. Resendiz-Lara, S. Azhdari, H. Gojzewski, A. H. Gröschel and F. R. Wurm, Chem. Sci., 2023, 14, 11273–11282 RSC.
- S. Schöttler, G. Becker, S. Winzen, T. Steinbach, K. Mohr, K. Landfester, V. Mailänder and F. R. Wurm, Nat. Nanotechnol., 2016, 11, 372–377 CrossRef.
- A. L. Mali, S. S. Priya and M. R. Rekha, J. Appl. Polym. Sci., 2021, 138 DOI:10.1002/app.51323.
- A. Ewe, O. Panchal, S. R. Pinnapireddy, U. Bakowsky, S. Przybylski, A. Temme and A. Aigner, Nanomedicine, 2017, 13, 209–218 CrossRef CAS PubMed.
- A. P. Pandey and K. K. Sawant, Mater. Sci. Eng., C, 2016, 68, 904–918 CrossRef CAS PubMed.
- F. Adams, C. M. Zimmermann, D. Baldassi, T. M. Pehl, P. Weingarten, I. Kachel, M. Kränzlein, D. C. Jürgens, P. Braubach, I. Alexopoulos, M. Wygrecka and O. M. Merkel, Small, 2024, 20 DOI:10.1002/smll.202308775.
- H. Zhang, K. Men, C. Pan, Y. Gao, J. Li, S. Lei, G. Zhu, R. Li, Y. Wei and X. Duan, Int. J. Nanomed., 2020, 15, 9875–9890 CrossRef CAS.
- P. Wu, X. Luo, H. Wu, F. Yu, K. Wang, M. Sun and D. Oupicky, Macromol. Biosci., 2018, 18 DOI:10.1002/mabi.201800234.
- T. Biswas and U. N. Dwivedi, Protoplasma, 2019, 256, 1463–1486 CrossRef CAS PubMed.
- S. Böttger and M. F. Melzig, Bioorg. Med. Chem., 2013, 21, 7118–7124 CrossRef PubMed.
- A. Weng, M. Thakur, B. Von Mallinckrodt, F. Beceren-Braun, R. Gilabert-Oriol, B. Wiesner, J. Eichhorst, S. Böttger, M. F. Melzig and H. Fuchs, J. Controlled Release, 2012, 164, 74–86 CrossRef CAS PubMed.
- M. A. Oberli, A. M. Reichmuth, J. R. Dorkin, M. J. Mitchell, O. S. Fenton, A. Jaklenec, D. G. Anderson, R. Langer and D. Blankschtein, Nano Lett., 2017, 17, 1326–1335 CrossRef CAS PubMed.
- A. Hasanzadeh, A. H. Vahabi, S. E. Hooshmand, E. S. Hosseini, B. K. Y. Azar, J. Kiani, S. Saeedi, A. Shahbazi, A. Rudra, M. R. Hamblin and M. Karimi, Nanotechnology, 2022, 33 DOI:10.1088/1361-6528/ac842d.
- H. Liu, Y. Wang, M. Wang, J. Xiao and Y. Cheng, Biomaterials, 2014, 35, 5407–5413 CrossRef CAS PubMed.
- J. Lv, H. Chang, Y. Wang, M. Wang, J. Xiao, Q. Zhang and Y. Cheng, J. Mater. Chem. B, 2015, 3, 642–650 RSC.
- E. Tan, J. Lv, J. Hu, W. Shen, H. Wang and Y. Cheng, J. Mater. Chem., 2018, 6, 7230–7238 RSC.
- J. Lv, B. He, J. Yu, Y. Wang, C. Wang, S. Zhang, H. Wang, J. Hu, Q. Zhang and Y. Cheng, Biomaterials, 2018, 182, 167–175 CrossRef CAS PubMed.
- H. Wang, J. Hu, X. Cai, J. Xiao and Y. Cheng, Polym. Chem., 2016, 7, 2319–2322 RSC.
- J. Lv and Y. Cheng, Chem. Soc. Rev., 2021, 50, 5435–5467 RSC.
- Z. Zhang, W. Shen, J. Ling, Y. Yan, J. Hu and Y. Cheng, Nat. Commun., 2018, 9 DOI:10.1038/s41467-018-03779-8.
- M. Cametti, B. Crousse, P. Metrangolo, R. Milani and G. Resnati, Chem. Soc. Rev., 2012, 41, 31–42 RSC.
- J. He, S. Xu and A. J. Mixson, Pharmaceutics, 2020, 12, 1–31 CrossRef PubMed.
- W. Y. Seow and Y. Y. Yang, Adv. Mater., 2009, 21, 86–90 CrossRef CAS.
- F. Heitz, M. C. Morris and G. Divita, Br. J. Pharmacol., 2009, 157, 195–206 CrossRef CAS PubMed.
- J. D. Torres-Vanegas, J. C. Cruz and L. H. Reyes, Pharmaceutics, 2021, 13 DOI:10.3390/pharmaceutics13030428.
- P. Ruzza, B. Biondi, A. Marchiani, N. Antolini and A. Calderan, Pharmaceuticals, 2010, 3, 1045–1062 CrossRef CAS PubMed.
- K. M. Stewart, K. L. Horton and S. O. Kelley, Org. Biomol. Chem., 2008, 6, 2242–2255 RSC.
- M. A. Gosselin, W. Guo and R. J. Lee, Bioconjugate Chem., 2001, 12, 989–994 CrossRef CAS PubMed.
- J. Liu, X. Jiang, L. Xu, X. Wang, W. E. Hennink and R. Zhuo, Bioconjugate Chem., 2010, 21, 1827–1835 CrossRef CAS PubMed.
- S. Bauhuber, R. Liebl, L. Tomasetti, R. Rachel, A. Goepferich and M. Breunig, J. Controlled Release, 2012, 162, 446–455 CrossRef CAS PubMed.
- Y. Lei, J. Wang, C. Xie, E. Wagner, W. Lu, Y. Li, X. Wei, J. Dong and M. Liu, J. Gene Med., 2013, 15, 291–305 CrossRef CAS PubMed.
- L. V. Christensen, C. W. Chang, J. K. Won, W. K. Sung, Z. Zhong, C. Lin, J. F. J. Engbersen and J. Feijen, Bioconjugate Chem., 2006, 17, 1233–1240 CrossRef CAS PubMed.
- J. Chen, C. Wu and D. Oupický, Biomacromolecules, 2009, 10, 2921–2927 CrossRef CAS PubMed.
- C. Wu, J. Li, Y. Zhu, J. Chen and D. Oupický, Biomaterials, 2013, 34, 8843–8850 CrossRef CAS PubMed.
- N. Zhang, Y. Wang, R. Wu, C. Xu, J. J. Nie, N. Zhao, B. Yu, Z. Liu and F. J. Xu, Small, 2019, 15 DOI:10.1002/smll.201904017.
- J. Xiang, X. Liu, Z. Zhou, D. Zhu, Q. Zhou, Y. Piao, L. Jiang, J. Tang, X. Liu and Y. Shen, ACS Appl. Mater. Interfaces, 2018, 10, 43352–43362 CrossRef CAS PubMed.
- D. Ma, H. Liu, P. Zhao, L. Ye, H. Zou, X. Zhao, H. Dai, X. Kong and P. Liu, ACS Appl. Mater. Interfaces, 2020, 12, 9032–9040 CrossRef CAS PubMed.
- B. Tyler, D. Gullotti, A. Mangraviti, T. Utsuki and H. Brem, Adv. Drug Delivery Rev., 2016, 107, 163–175 CrossRef CAS PubMed.
- S. Lanzalaco and E. Armelin, Gels, 2017, 3 Search PubMed.
- S. Y. Jeon, J. S. Park, H. N. Yang, D. G. Woo and K. H. Park, Biomaterials, 2012, 33, 4413–4423 CrossRef CAS PubMed.
- Y. C. Wang, Y. Y. Yuan, J. Z. Du, X. Z. Yang and J. Wang, Macromol. Biosci., 2009, 9, 1154–1164 CrossRef CAS PubMed.
- F. He, C. F. Wang, T. Jiang, B. Han and R. X. Zhuo, Biomacromolecules, 2010, 11, 3028–3035 CrossRef CAS PubMed.
- M. Gong, H. Liu, N. Sun, Y. Xie, F. Yan and L. Cai, Nanomedicine, 2020, 15, 711–723 CrossRef CAS PubMed.
- Z. H. Wang, Y. Zhu, M. Y. Chai, W. T. Yang and F. J. Xu, Biomaterials, 2012, 33, 1873–1883 CrossRef CAS PubMed.
- W. W. Hu, C. C. Yeh and C. W. Tsai, J. Mater. Chem. B, 2018, 6, 5781–5794 RSC.
- V. Bulmus, M. Woodward, L. Lin, N. Murthy, P. Stayton and A. Hoffman, in Journal of Controlled Release, Elsevier, 2003, vol. 93, pp. 105–120 Search PubMed.
- P. Erbacher, M. T. Bousser, J. Raimond, M. Monsigny, P. Midoux and A. C. Roche, Hum. Gene Ther., 1996, 7, 721–729 CrossRef CAS PubMed.
- A. V. Lopukhov, Z. Yang, M. J. Haney, T. K. Bronich, M. Sokolsky-Papkov, E. V. Batrakova, N. L. Klyachko and A. V. Kabanov, Macromol. Biosci., 2021, 21 DOI:10.1002/mabi.202000371.
- S. S. Diebold, M. Kursa, E. Wagner, M. Cotten and M. Zenke, J. Biol. Chem., 1999, 274, 19087–19094 CrossRef CAS PubMed.
- M. Elfinger, J. Geiger, G. Hasenpusch, S. Üzgün, N. Sieverling, M. K. Aneja, C. Maucksch and C. Rudolph, J. Controlled Release, 2009, 135, 234–241 CrossRef CAS PubMed.
- Y. Luo, X. Zhai, C. Ma, P. Sun, Z. Fu, W. Liu and J. Xu, J. Controlled Release, 2012, 162, 28–36 CrossRef CAS PubMed.
- C. Aranda, K. Urbiola, A. Méndez Ardoy, J. M. García Fernández, C. Ortiz Mellet and C. T. De Ilarduya, Eur. J. Pharm. Biopharm., 2013, 85, 390–397 CrossRef CAS PubMed.
- B. Liang, M. L. He, Z. P. Xiao, Y. Li, C. Y. Chan, H. F. Kung, X. T. Shuai and Y. Peng, Biochem. Biophys. Res. Commun., 2008, 367, 874–880 CrossRef CAS PubMed.
- Y. Rui, M. Varanasi, S. Mendes, H. M. Yamagata, D. R. Wilson and J. J. Green, Mol. Ther. – Nucleic Acids, 2020, 20, 661–672 CrossRef CAS PubMed.
- R. S. Burke and S. H. Pun, Bioconjugate Chem., 2010, 21, 140–150 CrossRef CAS PubMed.
- H. Wei, J. A. Pahang and S. H. Pun, Biomacromolecules, 2013, 14, 275–284 CrossRef CAS PubMed.
- T. Liu, W. Xue, B. Ke, M. Q. Xie and D. Ma, Biomaterials, 2014, 35, 3865–3872 CrossRef CAS PubMed.
- K. M. Xiu, J. J. Yang, N. N. Zhao, J. S. Li and F. J. Xu, Acta Biomater., 2013, 9, 4726–4733 CrossRef CAS PubMed.
- W. Chen, Y. Hong, T. Zhang, D. Kong, M. Zhang, Q. Zhang and C. Wang, Colloids Surf., B, 2019, 181, 721–727 CrossRef CAS PubMed.
- T. Wolf, T. Steinbach and F. R. Wurm, Macromolecules, 2015, 48, 3853–3863 CrossRef CAS.
- M. Elsabahy, S. Zhang, F. Zhang, Z. J. Deng, Y. H. Lim, H. Wang, P. Parsamian, P. T. Hammond and K. L. Wooley, Sci. Rep., 2013, 3 DOI:10.1038/srep03313.
- T. M. Sun, J. Z. Du, Y. D. Yao, C. Q. Mao, S. Dou, S. Y. Huang, P. Z. Zhang, K. W. Leong, E. W. Song and J. Wang, ACS Nano, 2011, 5, 1483–1494 CrossRef CAS PubMed.
- S. Zhang, J. Zou, M. Elsabahy, A. Karwa, A. Li, D. A. Moore, R. B. Dorshow and K. L. Wooley, Chem. Sci., 2013, 4, 2122–2126 RSC.
- J. Wang, P. C. Zhang, H. F. Lu, N. Ma, S. Wang, H. Q. Mao and K. W. Leong, J. Controlled Release, 2002, 83, 157–168 CrossRef CAS PubMed.
- A. Huwiler and U. Zangemeister-Wittke, Crit. Rev. Oncol. Hematol., 2007, 63, 150–159 CrossRef PubMed.
- Z. Liu, Z. Zhang, C. Zhou and Y. Jiao, Prog. Polym. Sci., 2010, 35, 1144–1162 CrossRef CAS.
- Y. Jiang, T. P. Lodge and T. M. Reineke, J. Am. Chem. Soc., 2018, 140, 11101–11111 CrossRef CAS PubMed.
- S. S. Parelkar, D. Chan-Seng and T. Emrick, Biomaterials, 2011, 32, 2432–2444 CrossRef CAS PubMed.
- J. O'Keeffe Ahern, A. Sigen, D. Zhou, Y. Gao, J. Lyu, Z. Meng, L. Cutlar, L. Pierucci and W. Wang, Biomacromolecules, 2018, 19, 1410–1415 CrossRef PubMed.
- J. Xu, Z. Li, Q. Fan, J. Lv, Y. Li and Y. Cheng, Adv. Healthcare Mater., 2018, 7 DOI:10.1002/adma.202104355.
- G. Candiani, M. Frigerio, F. Viani, C. Verpelli, C. Sala, L. Chiamenti, N. Zaffaroni, M. Folini, M. Sani, W. Panzeri and M. Zanda, ChemMedChem, 2007, 2, 292–296 CrossRef CAS PubMed.
- A. M. Doody, J. N. Korley, K. P. Dang, P. N. Zawaneh and D. Putnam, J. Controlled Release, 2006, 116, 227–237 CrossRef CAS PubMed.
- J. Lv and Y. Cheng, Chem. Soc. Rev., 2021, 50, 5435–5467 RSC.
- Y. P. Xiao, J. Zhang, Y. H. Liu, J. H. Zhang, Q. Y. Yu, Z. Huang and X. Q. Yu, Eur. J. Med. Chem., 2019, 162, 602–611 CrossRef CAS PubMed.
- M. Cametti, B. Crousse, P. Metrangolo, R. Milani and G. Resnati, Chem. Soc. Rev., 2012, 41, 31–42 RSC.
- L. Xue, Y. Yan, P. Kos, X. Chen and D. J. Siegwart, Drug Delivery Transl. Res., 2021, 11, 255–260 CrossRef CAS PubMed.
- G. Chen, K. Wang, Y. Wang, P. Wu, M. Sun and D. Oupický, Adv. Healthcare Mater., 2018, 7 DOI:10.1002/adhm.201700978.
- M. Wang, H. Liu, L. Li and Y. Cheng, Nat. Commun., 2015, 4 DOI:10.1038/ncomms4053.
- J. Lv, H. Chang, Y. Wang, M. Wang, J. Xiao, Q. Zhang and Y. Cheng, J. Mater. Chem. B, 2015, 3, 642–650 RSC.
- E. Tan, J. Lv, J. Hu, W. Shen, H. Wang and Y. Cheng, J. Mater. Chem. B, 2018, 6, 7230–7238 RSC.
- J. He, S. Xu and A. J. Mixson, Pharmaceutics, 2020, 12, 1–31 CrossRef PubMed.
- W. Y. Seow and Y. Y. Yang, Adv. Mater., 2009, 21, 86–90 CrossRef CAS.
- F. Heitz, M. C. Morris and G. Divita, Br. J. Pharmacol., 2009, 157, 195–206 CrossRef CAS PubMed.
- J. D. Torres-Vanegas, J. C. Cruz and L. H. Reyes, Pharmaceutics, 2021, 13, 428 CrossRef CAS PubMed.
- P. Ruzza, B. Biondi, A. Marchiani, N. Antolini and A. Calderan, Pharmaceuticals, 2010, 3, 1045–1062 CrossRef CAS PubMed.
- K. M. Stewart, K. L. Horton and S. O. Kelley, Org. Biomol. Chem., 2008, 6, 2242–2255 RSC.
- Z. Pan, X. Kang, Y. Zeng, W. Zhang, H. Peng, J. Wang, W. Huang, H. Wang, Y. Shen and Y. Huang, Polym. Chem., 2017, 8, 5275–5285 RSC.
- G. Francis, Z. Kerem, H. P. S. Makkar and K. Becker, Br. J. Nutr., 2002, 88, 587–605 CrossRef CAS PubMed.
- L. Zhou, M. Emenuga, S. Kumar, Z. Lamantia, M. Figueiredo and T. Emrick, Biomacromolecules, 2022, 23, 4029–4040 CrossRef CAS PubMed.
- I. Ullah, K. Muhammad, M. Akpanyung, A. Nejjari, A. L. Neve, J. Guo, Y. Feng and C. Shi, J. Mater. Chem. B, 2017, 5, 3253–3276 RSC.
- V. Bulmus, M. Woodward, L. Lin, N. Murthy, P. Stayton and A. Hoffman, in Journal of Controlled Release, Elsevier, 2003, vol. 93, pp. 105–120 Search PubMed.
- Y. Li, J. Ding, X. Xu, R. Shi, P. E. Saw, J. Wang, S. Chung, W. Li, B. M. Aljaeid, R. J. Lee, W. Tao, L. Teng, O. C. Farokhzad and J. Shi, Nano Lett., 2020, 20, 4857–4863 CrossRef CAS PubMed.
- X. Yang, Q. Tang, Y. Jiang, M. Zhang, M. Wang and L. Mao, J. Am. Chem. Soc., 2019, 141, 3782–3786 CrossRef CAS PubMed.
- F. Perche, S. Biswas, T. Wang, L. Zhu and V. P. Torchilin, Angew. Chem., Int. Ed., 2014, 53, 3362–3366 CrossRef CAS PubMed.
- L. Kang, B. Fan, P. Sun, W. Huang, M. Jin, Q. Wang and Z. Gao, Acta Biomater., 2016, 44, 341–354 CrossRef CAS PubMed.
- D. Neri and C. T. Supuran, Nat. Rev. Drug Discovery, 2011, 10, 767–777 CrossRef CAS PubMed.
- J. Li, L. Mo, C. H. Lu, T. Fu, H. H. Yang and W. Tan, Chem. Soc. Rev., 2016, 45, 1410–1431 RSC.
- M. Karimi, M. Eslami, P. Sahandi-Zangabad, F. Mirab, N. Farajisafiloo, Z. Shafaei, D. Ghosh, M. Bozorgomid, F. Dashkhaneh and M. R. Hamblin, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2016, 8, 696–716 CAS.
- Y. Wei, R. Liao, A. A. Mahmood, H. Xu and Q. Zhou, Acta Biomater., 2017, 55, 194–203 CrossRef CAS PubMed.
- C. Liu, G. Chern and F. Hsu, Hepatology, 2018, 67, 899–913 CrossRef CAS PubMed.
- J. Huang, C. Lin, J. Fang, X. Li, J. Wang, S. Deng, S. Zhang, W. Su, X. Feng, B. Chen, D. Cheng and X. Shuai, ACS Appl. Mater. Interfaces, 2018, 10, 28471–28482 CrossRef CAS PubMed.
- X. Guan, Z. Guo, L. Lin, J. Chen, H. Tian and X. Chen, Nano Lett., 2016, 16, 6823–6831 CrossRef CAS PubMed.
- R. Tang, W. Ji and C. Wang, Polymer, 2011, 52, 921–932 CrossRef CAS PubMed.
- Y. Qi, H. Song, H. Xiao, G. Cheng, B. Yu and F. J. Xu, Small, 2018, 14 DOI:10.1002/smll.201803061.
- G. Saravanakumar, J. Kim and W. J. Kim, Adv. Sci., 2017, 4 Search PubMed.
- Q. Xu, C. He, C. Xiao and X. Chen, Macromol. Biosci., 2016, 16, 635–646 CrossRef CAS PubMed.
- J. H. Zhang, W. J. Wang, J. Zhang, Y. P. Xiao, Y. H. Liu and X. Q. Yu, Eur. J. Med. Chem., 2019, 182, 2–10 Search PubMed.
- S. Matsumoto, R. J. Christie, N. Nishiyama, K. Miyata, A. Ishii, M. Oba, H. Koyama, Y. Yamasaki and K. Kataoka, Biomacromolecules, 2009, 10, 119–127 CrossRef CAS PubMed.
- C. Lin, Z. Zhong, M. C. Lok, X. Jiang, W. E. Hennink, J. Feijen and J. F. J. Engbersen, Bioconjugate Chem., 2007, 18, 138–145 CrossRef CAS PubMed.
- E. K. Bang, M. Lista, G. Sforazzini, N. Sakai and S. Matile, Chem. Sci., 2012, 3, 1752–1763 RSC.
- P. Laskar and C. Dufès, Nanoscale Adv., 2021, 3, 6007–6026 RSC.
- R. Dong, Y. Zhou, X. Huang, X. Zhu, Y. Lu and J. Shen, Adv. Mater., 2015, 27, 498–526 CrossRef CAS PubMed.
- G. Candiani, D. Pezzoli, L. Ciani, R. Chiesa and S. Ristori, PLoS One, 2010, 5 DOI:10.1371/journal.pone.0013430.
- Y. Zhang, L. Lin, L. Liu, F. Liu, A. Maruyama, H. Tian and X. Chen, Carbohydr. Polym., 2018, 201, 246–256 CrossRef CAS PubMed.
- D. M. Beaupre and R. G. Weiss, Molecules, 2021, 26 Search PubMed.
- Q. Liu, R. C. Su, W. J. Yi and Z. G. Zhao, Molecules, 2017, 22 DOI:10.3390/molecules22040566.
- F. Q. Schafer and G. R. Buettner, Free Radical Biol. Med., 2001, 30, 1191–1212 CrossRef CAS PubMed.
- D. Jere, J. E. Kim, R. Arote, H. L. Jiang, Y. K. Kim, Y. J. Choi, C. H. Yun, M. H. Cho and C. S. Cho, Biomaterials, 2009, 30, 1635–1647 CrossRef CAS PubMed.
- Y. Fu, Y. Sun, M. Chen, W. Xing, Y. Xu, X. Qian and W. Zhu, Biomacromolecules, 2022, 23, 1251–1258 CrossRef CAS PubMed.
- S. Ma, W. Song, Y. Xu, X. Si, S. Lv, Y. Zhang, Z. Tang and X. Chen, Nano Lett., 2020, 20, 2514–2521 CrossRef CAS PubMed.
- X. Liu, J. M. Holzwarth and P. X. Ma, Macromol. Biosci., 2012, 12, 911–919 CrossRef CAS PubMed.
- N. L. Benner, R. L. McClellan, C. R. Turlington, O. A. W. Haabeth, R. M. Waymouth and P. A. Wender, J. Am. Chem. Soc., 2019, 141, 8416–8421 CrossRef CAS PubMed.
- Z. Wang, C. Huang, H. Lv, M. Zhang and X. Li, PLoS One, 2020, 15, 1–19 Search PubMed.
- Y. Liu, Y. Li, D. Keskin and L. Shi, Adv. Healthcare Mater., 2019, 8 Search PubMed.
- C. K. Williams, Chem. Soc. Rev., 2007, 36, 1573–1580 RSC.
- J. Pretula, S. Slomkowski and S. Penczek, Adv. Drug Delivery Rev., 2016, 107, 3–16 CrossRef CAS PubMed.
- R. P. Brannigan and A. P. Dove, Biomater. Sci., 2017, 5, 9–21 RSC.
- N. Y. Steinman, R. L. Starr, S. D. Brucks, C. Belay, R. Meir, J. Golenser, L. M. Campos and A. J. Domb, Macromolecules, 2019, 52, 3543–3550 CrossRef CAS.
- X. B. Xiong, H. Uludaǧ and A. Lavasanifar, Biomaterials, 2009, 30, 242–253 CrossRef CAS PubMed.
- W. C. Hung, M. Da Shau, H. C. Kao, M. F. Shih and J. Y. Cherng, J. Controlled Release, 2009, 133, 68–76 CrossRef CAS PubMed.
- X. Y. Liu, W. Y. Ho, W. J. Hung and M. Da Shau, Biomaterials, 2009, 30, 6665–6673 CrossRef CAS PubMed.
- Y. P. Yang, Y. Chien, G. Y. Chiou, J. Y. Cherng, M. L. Wang, W. L. Lo, Y. L. Chang, P. I. Huang, Y. W. Chen, Y. H. Shih, M. T. Chen and S. H. Chiou, Biomaterials, 2012, 33, 1462–1476 CrossRef CAS PubMed.
- G. Y. Chiou, J. Y. Cherng, H. S. Hsu, M. L. Wang, C. M. Tsai, K. H. Lu, Y. Chien, S. C. Hung, Y. W. Chen, C. I. Wong, L. M. Tseng, P. I. Huang, C. C. Yu, W. H. Hsu and S. H. Chiou, J. Controlled Release, 2012, 159, 240–250 CrossRef CAS PubMed.
- W. L. Lo, Y. Chien, G. Y. Chiou, L. M. Tseng, H. S. Hsu, Y. L. Chang, K. H. Lu, C. S. Chien, M. L. Wang, Y. W. Chen, P. I. Huang, F. W. Hu, C. C. Yu, P. Y. Chu and S. H. Chiou, Biomaterials, 2012, 33, 3693–3709 CrossRef CAS PubMed.
- W. Chen, F. Meng, R. Cheng, C. Deng, J. Feijen and Z. Zhong, J. Controlled Release, 2014, 190, 398–414 CrossRef CAS PubMed.
- C. F. Wang, Y. X. Lin, T. Jiang, F. He and R. X. Zhuo, Biomaterials, 2009, 30, 4824–4832 CrossRef CAS PubMed.
- F. He, C. F. Wang, T. Jiang, B. Han and R. X. Zhuo, Biomacromolecules, 2010, 11, 3028–3035 CrossRef CAS PubMed.
- Z. H. Wang, W. B. Li, J. Ma, G. P. Tang, W. T. Yang and F. J. Xu, Macromolecules, 2011, 44, 230–239 CrossRef CAS.
- M. Gong, H. Liu, N. Sun, Y. Xie, F. Yan and L. Cai, Nanomedicine, 2020, 15, 711–723 CrossRef CAS PubMed.
- Z. H. Wang, Y. Zhu, M. Y. Chai, W. T. Yang and F. J. Xu, Biomaterials, 2012, 33, 1873–1883 CrossRef CAS PubMed.
- S. A. Dilliard and D. J. Siegwart, Nat. Rev. Mater., 2023, 8, 282–300 CrossRef CAS PubMed.
- R. C. Steffens and E. Wagner, Pharm. Res., 2022, 40, 47–76 CrossRef PubMed.
- K. Paunovska, D. Loughrey and J. E. Dahlman, Nat. Rev. Genet., 2022, 23, 265–280 CrossRef CAS PubMed.
- M. Buschle, M. Cotten, H. Kirlappos, K. Mechtler, G. Schaffner, W. Zauner, M. L. Birnstiel and E. Wagner, Hum. Gene Ther., 1995, 6, 753–761 CrossRef CAS PubMed.
- D. Witzigmann, L. Quagliata, S. H. Schenk, C. Quintavalle, L. M. Terracciano and J. Huwyler, Hepatol. Res., 2016, 46, 686–696 CrossRef CAS PubMed.
- C. Plank, K. Zatloukal, M. Gotten, K. Mechtler and E. Wagner, Gene Transfer into Hepatocytes Using Asialoglycoprotein Receptor Mediated Endocytosis of DNA Complexed with an Artificial Tetra-Antennary Galactose Ligand, 1992, vol. 3 Search PubMed.
- M. Elfinger, C. Maucksch and C. Rudolph, Biomaterials, 2007, 28, 3448–3455 CrossRef CAS PubMed.
- S. Li, Y. Tan, E. Viroonchatapan, B. R. Pitt and L. Huang, Targeted gene delivery to pulmonary endothelium by anti-PECAM antibody, 2000 Search PubMed.
- J. Geiger, M. K. Aneja, G. Hasenpusch, G. Yüksekdag, G. Kummerlöwe, B. Luy, T. Romer, U. Rothbauer and C. Rudolph, Biomaterials, 2010, 31, 2903–2911 CrossRef CAS PubMed.
- W. J. Kollen, F. M. Schembri, G. J. Gerwig, J. F. Vliegenthart, M. C. Glick and T. F. Scanlin, Am. J. Respir. Cell Mol. Biol., 1999, 20, 1081–1086 CrossRef CAS PubMed.
- W. J. Kollen, A. E. Mulberg, X. Wei, M. Sugita, V. Raghuram, J. Wang, J. K. Foskett, M. C. Glick and T. F. Scanlin, Hum. Gene Ther., 1999, 10, 615–622 CrossRef CAS PubMed.
- W. J. Kollen, P. Midoux, P. Erbacher, A. Yip, A. C. Roche, M. Monsigny, M. C. Glick and T. F. Scanlin, Hum. Gene Ther., 1996, 7, 1577–1586 CrossRef CAS PubMed.
- J. Chen, X. Gao, K. Hu, Z. Pang, J. Cai, J. Li, H. Wu and X. Jiang, Biochem. Biophys. Res. Commun., 2008, 375, 378–383 CrossRef CAS PubMed.
- Y. Xie, N. H. Kim, V. Nadithe, D. Schalk, A. Thakur, A. Kılıç, L. G. Lum, D. J. P. Bassett and O. M. Merkel, J. Controlled Release, 2016, 229, 120–129 CrossRef CAS PubMed.
- R. Kandil, Y. Xie, R. Heermann, L. Isert, K. Jung, A. Mehta and O. M. Merkel, Adv. Ther., 2019, 2 DOI:10.1002/adtp.201900047.
- A. D'Souza, S. Nozohouri, B. S. Bleier and M. M. Amiji, Pharm. Res., 2023, 40, 77–105 CrossRef PubMed.
- R.-Q. Huang, Y.-H. Qu, W.-L. Ke, J.-H. Zhu, Y.-Y. Pei and C. Jiang, FASEB J., 2007, 21, 1117–1125 CrossRef CAS PubMed.
- R. Huang, W. Ke, Y. Liu, C. Jiang and Y. Pei, Biomaterials, 2008, 29, 238–246 CrossRef CAS PubMed.
- S. Somani, D. R. Blatchford, O. Millington, M. L. Stevenson and C. Dufès, J. Controlled Release, 2014, 188, 78–86 CrossRef CAS PubMed.
- S. Somani, G. Robb, B. S. Pickard and C. Dufès, J. Controlled Release, 2015, 217, 235–242 CrossRef CAS PubMed.
- F. Gosselet, R. A. Loiola, A. Roig, A. Rosell and M. Culot, Neurochem. Int., 2021, 144 DOI:10.1016/j.neuint.2020.104952.
- T. Benli-Hoppe, Ş. G. Öztürk, Ö. Öztürk, S. Berger, E. Wagner and M. Yazdi, Macromol. Rapid Commun., 2022, 43, 1–12 CrossRef PubMed.
- W. Ke, K. Shao, R. Huang, L. Han, Y. Liu, J. Li, Y. Kuang, L. Ye, J. Lou and C. Jiang, Biomaterials, 2009, 30, 6976–6985 CrossRef CAS PubMed.
- Y. Liu, X. He, Y. Kuang, S. An, C. Wang, Y. Guo, H. Ma, J. Lou and C. Jiang, Mol. Pharm., 2014, 11, 3330–3341 CrossRef CAS PubMed.
- S. Gottschalk, S. Jude, G. Therapy, S. Gottschalk, R. J. Cristiano, L. C. Smith and S. L. C. Woo’, Folate receptor mediated DNA delivery into tumor cells: Potosomal disruption results in enhanced gene expression Fola te recep for media ted DNA delivery into tumor cells: potosomal disruption results in enhanced gene expression, 1994, vol. 1 Search PubMed.
- C. Wang, W. He, F. Wang, H. Yong, T. Bo, D. Yao, Y. Zhao, C. Pan, Q. Cao, S. Zhang and M. Li, J. Nanobiotechnol., 2024, 22 CAS.
- N. Hartl, D. C. Jürgens, S. Carneiro, A. C. König, X. Xiao, R. Liu, S. M. Hauck and O. M. Merkel, Int. J. Pharm., 2023, 643 DOI:10.1016/j.ijpharm.2023.123257.
- S. Mirón-Barroso, J. S. Correia, A. E. Frampton, M. P. Lythgoe, J. Clark, L. Tookman, S. Ottaviani, L. Castellano, A. E. Porter, T. K. Georgiou and J. Krell, Noncoding RNAs, 2022, 8 Search PubMed.
- M. Neu, D. Fischer and T. Kissel, J. Gene Med., 2005, 7, 992–1009 CrossRef CAS PubMed.
- L. Parhamifar, A. K. Larsen, A. C. Hunter, T. L. Andresen and S. M. Moghimi, Soft Matter, 2010, 6, 4001–4009 RSC.
- Y. Rui, M. Varanasi, S. Mendes, H. M. Yamagata, D. R. Wilson and J. J. Green, Mol. Ther. – Nucleic Acids, 2020, 20, 661–672 CrossRef CAS PubMed.
- R. L. Xie, Y. J. Jang, L. Xing, B. F. Zhang, F. Z. Wang, P. F. Cui, M. H. Cho and H. L. Jiang, Int. J. Pharm., 2015, 478, 19–30 CrossRef CAS PubMed.
- X. Cai, R. Dou, C. Guo, J. Tang, X. Li, J. Chen and J. Zhang, Pharmaceutics, 2023, 15 Search PubMed.
- M. Szewczyk-Łagodzińska, A. Plichta, M. Dębowski, S. Kowalczyk, A. Iuliano and Z. Florjańczyk, Polymers, 2023, 15 Search PubMed.
- X. Liao, G. Walden, N. D. Falcon, S. Donell, M. J. Raxworthy, M. Wormstone, G. P. Riley and A. Saeed, Eur. Polym. J., 2017, 87, 458–467 CrossRef CAS.
- T. K. Georgiou, M. Vamvakaki, C. S. Patrickios, E. N. Yamasaki and L. A. Phylactou, Biomacromolecules, 2004, 5, 2221–2229 CrossRef CAS PubMed.
- F. J. Xu and W. T. Yang, Prog. Polym. Sci., 2011, 36, 1099–1131 CrossRef CAS.
- S. Srinivasachari, K. M. Fichter and T. M. Reineke, J. Am. Chem. Soc., 2008, 130, 4618–4627 CrossRef CAS PubMed.
- B. Tian and J. Liu, New J. Chem., 2020, 44, 9137–9148 RSC.
- W. Wu, W. Wang and J. Li, Prog. Polym. Sci., 2015, 46, 55–85 CrossRef CAS.
- J. M. Ren, T. G. McKenzie, Q. Fu, E. H. H. Wong, J. Xu, Z. An, S. Shanmugam, T. P. Davis, C. Boyer and G. G. Qiao, Chem. Rev., 2016, 116, 6743–6836 CrossRef CAS PubMed.
- S. Plesselova, P. Garcia-Cerezo, V. Blanco, F. J. Reche-Perez, F. Hernandez-Mateo, F. Santoyo-Gonzalez, M. D. Giron-Gonzalez and R. Salto-Gonzalez, J. Med. Chem., 2021, 64, 12245–12260 CrossRef CAS PubMed.
- T. Liu, W. Xue, B. Ke, M. Q. Xie and D. Ma, Biomaterials, 2014, 35, 3865–3872 CrossRef CAS PubMed.
- K. M. Xiu, J. J. Yang, N. N. Zhao, J. S. Li and F. J. Xu, Acta Biomater., 2013, 9, 4726–4733 CrossRef CAS PubMed.
- W. Chen, Y. Hong, T. Zhang, D. Kong, M. Zhang, Q. Zhang and C. Wang, Colloids Surf., B, 2019, 181, 721–727 CrossRef CAS PubMed.
- J. Shi, J. L. Choi, B. Chou, R. N. Johnson, J. G. Schellinger and S. H. Pun, ACS Nano, 2013, 7, 10612–10620 CrossRef CAS.
- T. G. Floyd, J. I. Song, A. Hapeshi, S. Laroque, M. Hartlieb and S. Perrier, J. Mater. Chem. B, 2020, 10, 3696–3704 RSC.
- R. S. Burke and S. H. Pun, Bioconjugate Chem., 2010, 21, 140–150 CrossRef CAS PubMed.
- H. Wei, J. A. Pahang and S. H. Pun, Biomacromolecules, 2013, 14, 275–284 CrossRef CAS.
- R. B. Breitenkamp and T. Emrick, Biomacromolecules, 2008, 9, 2495–2500 CrossRef CAS.
- A. F. Ghobadi, R. Letteri, S. S. Parelkar, Y. Zhao, D. Chan-Seng, T. Emrick and A. Jayaraman, Biomacromolecules, 2016, 17, 546–557 CrossRef CAS PubMed.
- J. Witten, Y. Hu, R. Langer and D. G. Anderson, Proc. Natl. Acad. Sci. U. S. A., 2021, 121 DOI:10.1073/pnas.2307798120.
- A. I. S. van den Berg, C. O. Yun, R. M. Schiffelers and W. E. Hennink, J. Controlled Release, 2021, 331, 121–141 CrossRef CAS PubMed.
- J. Casper, S. H. Schenk, E. Parhizkar, P. Detampel, A. Dehshahri and J. Huwyler, J. Controlled Release, 2023, 362, 667–691 CrossRef CAS PubMed.
- G. Protopapa, N. Bono, R. Visone, F. D'Alessandro, M. Rasponi and G. Candiani, Lab Chip, 2022, 23, 136–145 RSC.
- E. Giupponi, R. Visone, P. Occhetta, F. Colombo, M. Rasponi and G. Candiani, Biotechnol. Bioeng., 2018, 115, 775–784 CrossRef CAS PubMed.
- J. Kim, I. Hwang, D. Britain, T. D. Chung, Y. Sun and D. H. Kim, Lab Chip, 2011, 11, 3941–3948 RSC.
- H. Parhiz, E. N. Atochina-Vasserman and D. Weissman, Lancet, 2024, 403, 1192–1204 CrossRef CAS.
- A. J. Barbier, A. Y. Jiang, P. Zhang, R. Wooster and D. G. Anderson, Nat. Biotechnol., 2022, 40, 840–854 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |