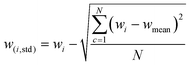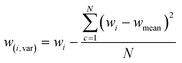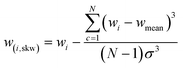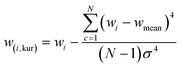Open Access Article
Open Access ArticleEnhancing glucose classification in continuous flow hydrothermal biomass liquefaction streams through generative AI and IR spectroscopy†
Silviu Florin
Acaru
 *a,
Rosnah
Abdullah
*a,
Rosnah
Abdullah
 b,
Daphne Teck Ching
Lai
b,
Daphne Teck Ching
Lai
 c and
Ren Chong
Lim
c and
Ren Chong
Lim
 *a
*a
aCentre for Advanced Material and Energy Sciences (CAMES), Universiti Brunei Darussalam, Jalan Tungku Link, Gadong BE1410, Brunei Darussalam. E-mail: s.f.acaru@outlook.com; renchong.lim@ubd.edu.bn
bFaculty of Science (FOS), Universiti Brunei Darussalam, Jalan Tungku Link, Gadong BE1410, Brunei Darussalam
cSchool of Digital Science (SDS), Universiti Brunei Darussalam, Jalan Tungku Link, Gadong BE1410, Brunei Darussalam
First published on 18th July 2023
Abstract
Energy from fossil fuels is forecasted to contribute to 28% of the energy demand by 2050. Shifting to renewable, green energy is desirable to mitigate the adverse effects on the climate posed by resultant gases. Continuous flow hydrothermal liquefaction holds promise to convert biomass into renewable energy. However, sustainable conversion of biomass feedstocks remains a considerable challenge, and more process optimization studies are necessary to achieve positive net energy ratios (NERs). To fast-track this process development, we investigated the integration of Fourier transform infrared spectroscopy (FTIR) for data collection coupled with a support vector machine classifier (SVC). We trained the model on data labeled after the analysis of the aqueous stream by high-performance liquid chromatography (HPLC). Multiple test data, such as liquified wood and cotton, and dissolved glucose, were used to classify the aqueous streams. The results showed that fused original data achieves 84% accuracy. The accuracy increased to 93% after merging synthetic data from generative adversarial networks (GANs) and hand-crafted statistical features. The effect of Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) on accuracy was also studied. We noticed that UMAP increases accuracy on some variations of the datasets, but it does not exceed the highest reported value. Shapely Additive Explanations (SHAP) were used to investigate the contribution of the top 20 features. We discovered that features representative of glucose contribute positively to the model's performance, whereas those found in water have a negative influence.
Introduction
Hydrothermal liquefaction (HTL) is a thermal conversion method that can decompose biomass such as sewage sludge, algae, oils, or lignocellulosic materials into biofuels. Achieving sustainability in HTL relies on the efficient energy conversion, ensuring a net energy gain that surpasses the energy input with its corresponding energy output. Extensive research has been conducted on various configurations, integrated technologies, and reporting metrics to address this matter. For example, the microwave-assisted batch HTL of lignocellulosic biomass revealed that the energy ratio of biocrude increases by 0.4 value points at a retention time of 60 minutes and a temperature of 240 °C.1 Recycling the aqueous phase in a catalytic HTL can impact the energy consumption within a 38% to 80% range, under varied temperature conditions.2 Sequential HTL treatments as alternative methods for biomass pretreatment showed increase glucose yields of 0.5% and 3% from poplar wood chips at temperatures of 140 °C and 180 °C, respectively.3 Effects of conventional batch HTL on a low-lipid marine species demonstrated a gradual increase in energy recovery for carbohydrates over time. After 10, 20, and 30 minutes of retention time at a temperature of 350 °C, the recovery rates increase from 10%, to 15%, and 29%, respectively.4 Starch as a feedstock showed a recovery rate of 25%, while cellulose yielded 23%. The incorporation of a catalyst such as cobalt and molybdenum doubles the energy recovery values for both feedstocks.5 A multi-cycle HTL showed that the initial energy recovery rate can increase from 49% up to 65% in the span of three cycles, while the use of catalysts under these conditions showed to lower the energy recovery rate to 55%.6 More recently, reaction atmosphere consisting of potassium hydroxide and hydrogen showed bio-oil yields up to 35 wt% in batch reactors.7Despite considerable progress achieved through different configurations, the sustainability of HTL systems is still not favorable for adhering to positive conversion principles. Continuous flow HTL emerges as a more appealing option over the batch and semi-continuous types due to its ability to generate fuel materials in large quantities and extract desired compounds while controlling the biomass retention periods.8 During the conversion process in continuous flow HTL, two primary by-products are generated: a solid residue and an aqueous phase. The aqueous phase is rich in fermentable sugars and other valuable compounds. The sugars can be further exploited for their calorific properties or enhanced through fermentation to generate high-yield liquid fuels, a form of renewable energy.9
The sustainability of the continuous flow HTL system is deemed favourable when the Net Energy Ratios (NERs) exceed 100%. The NER serves as a measure of the energy yield of a compound relative to the energy input into the system. Unlike other reported metrics such as energy recovery, which solely consider the energy content of the resulting fuel or bio-oil obtained, NER offers a more comprehensive analysis by taking into account the total energy efficiency of the HTL system.
A preliminary study focusing on the conversion of pre-treated wood waste residues has demonstrated that continuous flow HTL can achieve glucose NER values as high as 63%.10 To enhance the optimization of biomass conversion in continuous flow HTL, additional studies are required to refine parameters optimization, biomass load-to-weight ratio, and pre-treatment methods. Nevertheless, the rapid optimization of biomass conversion in continuous flow HTL encounters two primary challenges.
The first challenge arises from traditional optimization studies, which necessitate significant resources such as consumables, energy, time, and skilled labour. For instance, high-performance liquid chromatography (HPLC), an offline analysis technique used to determine compound concentrations in the aqueous phase, provides highly reliable data. However, the HPLC analysis entails a series of labour-intensive steps, including sample preparation, instrument qualification, compound identification, and quantification.
The second challenge lies in the intricate nature of HPLC analysis, which hampers the swift adjustment and control of HTL parameters during testing. To address these limitations, alternative methods that provide fast, cost-effective, and inline measurements can be employed. Fourier transform infrared spectroscopy (FTIR) is one such technique that enables simultaneous analysis of complex mixtures. It offers qualitative and quantitative information of sufficient accuracy, making it a valuable complement to overcome these challenges. This technique can analyse complex mixtures simultaneously, revealing adequate qualitative and quantitative information.11 With regards to aqueous solutions, FTIR has applications in several fields, such as diabetes monitoring,12 food additives,13 allergens14 and bio-hybrid fuel cells.15 Compounds of interest resulting from conversion processes have also been analysed, such as the sugar content in enzymatic hydrolysis of alkali-pretreated biomasses,16 the quantification of glucose in aqueous solutions,17 and quantification of aqueous phases (bio-crudes) derived from HTL.18 Additionally, apart from the analysis hindrance, the sheer number of experimental runs to reach conclusive results also slows down the process optimization.
Research applying machine learning (ML) algorithms to solve problems associated with energy studies is increasingly prevalent, ranging from material design models, discovery of unknown compounds and acceleration of innovation development such as high-performance fast charging batteries.19,20 The increasing importance of incorporating these concepts into hydrothermal liquefaction yields cannot be overstated. However, ML algorithms generally perform well when trained on large datasets. To complement the lack of data in determining the best HTL parameters, researchers resolve to compiling data from various published literature.21,22 In the case of continuous flow HTL, this approach is not feasible for two reasons:
a. there are not enough studies that published results using a similar HTL setup, and
b. the parameters and outputs are specific to the level of control and handling of the biomass.
Acknowledging the inadequacy of assuming uniform handling of all experiments, deep learning (DL), a subfield of ML, offers powerful algorithms that effectively tackle the challenges posed by limited data availability and expedite the optimization process. These DL algorithms play a crucial role in enhancing learning capabilities and facilitating more efficient decision-making.
Among the notable techniques in DL, generative adversarial networks (GANs) stand out as a preeminent approach for augmenting data from real-world examples, particularly in low data scenarios. GANs have demonstrated their effectiveness across various domains, including the design of materials models,19 generation of realistic medical images,23 object detection,24 augmentation of sensory signals,25 and improvement of Raman spectroscopy data.26 The latter is analogous to the infrared spectra obtained through FTIR.
Nonetheless, correct identification of the molecules in aqueous solution by FTIR is challenging due to the contribution of water molecules to the absorption spectrum. Absorption peaks of chemical bonds under aqueous mid-infrared radiation are broad, spreading across several wavelengths.11 Solutions such as feature engineering using statistical values has shown to capture the interconnection of movements by depth sensors.27 Similarly, hand-crafted statistical features could also be applicable to the vibrational intensity across wavelengths and amplify the response.28 However, FTIR spectrums have regions that are not significantly important in explaining the presence of a compound and with the generation of statistical features, insignificant values are introduced for each sample. Training a model on irrelevant data can have a negative impact on model's performance. Dimensionality reduction techniques, such as the Uniform Manifold Approximation for Projection (UMAP) algorithm can be used to improve a model's performance. UMAP selects the essential features using nearest neighbours to construct the simplicial set.29 The question remains whether the final ML model is reliable, and to confirm, one needs to ensure that the contribution of the significant features are the determining ones. Shapley Additive Explanations (SHAP) can be used to interpret each value of the features and understand the respective contribution of the vibrational spectra wavelengths.30
Therefore, the aim of this study is to implement a ML model into a continuous flow HTL system that could rapidly classify samples with high accuracy and confidence during biomass conversion into biofuel materials. The study's objectives are as follows:
1. Investigate the suitability of GANs in synthetic data generation from FTIR spectrums to increase dataset size for improving ML classification performance.
2. Enhance the model's performance using hand-crafted statistical features and a dimensionality reduction technique.
3. Verify whether the features with significant importance in glucose compounds are contributing positively to the model's performance.
The novel proposed framework will accelerate glucose recognition in the aqueous phase from the continuous flow HTL conversion process when the level is above a set threshold. The framework involves numerical data collected from three different experiments. The first dataset is derived from wood (W) waste and it represents the minimum viable real data of the lignocellulosic biomass conversion process. The second dataset is derived from conversion of cotton (C), which is a cleaner lignocellulosic biomass representative. The last dataset is attained from dissolved glucose (DG) with a high purity content. The dissolved glucose dataset is meant to enforce the model into training with more samples, representative of the target material.
Fourier transform infrared spectroscopy
FTIR spectroscopy is an increasingly versatile and rapid analytical technique which provides high information content in the form of vibrational spectra. This technique characterizes samples in various states: e.g., solid, gas, and aqueous environments and it has been proven successful in both in-line and offline process monitoring.31 Accessories can enhance the instrument's usability in different modes, providing micro and macro imagining, attenuated total reflectance (ATR), transflection, and transmission methods. Analytically, ATR-FTIR captures sample information at depths between 0.5–5 μm, which is sufficient for measuring aqueous solutions where water penetration in transmission modes reaches 6 μm at most.32 The sensory output can be delivered in image or comma separated values (CSV) formats. In this study, CSV is used and the contained data is processed into a tabular type.Generative adversarial networks
GANs are deep-learning-based generative models using Neural Networks.33 They have capacity to learn an intricate high-dimensional probability distribution and to produce high quality samples from the different data (e.g., images, text, tables). The principles behind GANs involve a generator (G) and a discriminator (D) model. The two models are in an ongoing competition governed by the min-max GAN loss (eqn (1)). G aims to minimize the function (V), whereas D strives to maximize it.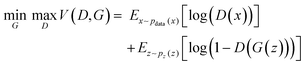 | (1) |
In other words, the generator model is responsible for generating new data samples from a given dataset. In contrast, the discriminator model acts as a classifier and tries to distinguish whether the new data sample is real or fake by comparing the training and fake data.
Numerous variations have been proposed over the traditional GANs. For example, the Wasserstein model (WGAN) improved the training stability by introducing the Earth-Mover distance (or Wasserstein-1) to the loss function.34 Still, the model experienced difficulties in generating accurate samples due to weight clipping. As a remedy, improvements such as gradient penalty (WGAN-GP) to the original critic loss showed promising results.35 While some architectures focused on generating new image variations, others concentrated on table data types (tabular). The implicit joint distribution of columns, which is the probability of two variables happening together, can be learned from the real data. Synthetic data can be produced from the resulted distribution. Algorithms such as tabular GAN (TGAN) and conditional tabular GAN (CTGAN) which are based on recurrent networks, outperformed previous statistical ways of augmenting tabular data (e.g., classification, regression trees, and Bayesian networks).36 Table-GAN, which is based on convolutional neural networks, is another case model that generates synthetic valuable tabular data.37 The interest in synthetic data and the proven capability of this new form of data augmentation is in the incipient stages. Continuous improvements are being reported at very fast pace but no studies looked at generating synthetic data using near-infrared spectrums captured by ATR-FTIR. In this study, the standard structure of GAN as outlined in the work reported by ref. 33 is adopted. Detailed implementation instructions can be found within the Data processing and augmentation section.
Experimental
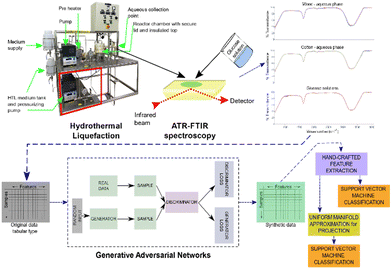
The overall framework of the study is detailed in Fig. 1. The study was initiated with the collection of experimental samples. In this step, the wood and cotton were sequentially decomposed into biofuel materials. A range of HTL parameters were employed under subcritical water conditions, with specific details provided in Tables A I and A II in the ESI.† Regular collection of aqueous samples was performed, and they were then subjected to the ATR-FTIR instrument at ambient temperature, where their characteristic transmittance was recorded. The purified samples were subsequently passed through an HPLC instrument to analyse the presence or absence of biofuel materials. The HPLC instrument separated compounds by passing the solution through a separation column, and the detector at the end of the column accurately identified and quantified each compound with a high degree of accuracy. The results obtained from the HPLC analysis were used to encode labels for the spectroscopy data acquired from ATR-FTIR measurements.Experimental machine set up and biomass samples
The HTL system and the biomass samples utilized for data generation were previously described in ref. 10. For the experimental setup, both cotton and dissolved glucose were employed to create comparable HTL aqueous streams. More detailed information can be found in the Materials and methods section provided in the ESI.†Data acquisition for model building
Data processing and augmentation
The augmentation of synthetic data involved the utilization of several open-source libraries, including Numpy, scikit-learn, os, Matplotlib, and Keras. These libraries provide crucial functionalities for manipulating, pre-processing, visualizing, and constructing neural network models with the data.Data pre-processing encompassed the manipulation necessary to adhere to a matrix structure. For instance, in the wood dataset, the dimensions were established as 24 rows and 900 columns, the cotton dataset consisted of 39 rows and 900 columns, and the dissolved glucose dataset was shaped into 40 rows and 900 columns. Each row in these datasets represents an analyzed sample of the aqueous fluid conducted through the FTIR analysis, with the features designated as wavelengths. Each sample was labeled according to the glucose concentration determined by the HPLC analysis.
To prepare the datasets for augmentation, they were individually loaded and subjected to further processing steps. These steps involved removing irrelevant columns, scaling the data using the Min-Max scaling technique, and dividing it into feature and label components.
The configuration of the GAN algorithm employed a multilayer perceptron architecture. Within the code (available under this link: https://github.com/silviu20/GAN_IR_Spectroscopy.git), various essential functions were specified to facilitate the augmentation process. One such function was “generate_latent_points(latent_dim, n_samples),” which generates random points (latent space vectors) by sampling from a standard normal distribution. These points serve as input for the generator model. Another crucial function is “generate_fake_samples(generator, latent_dim, n_samples),” which generates counterfeit samples by feeding randomly generated latent points into the generator model. The resulting samples are labeled as “fake” (y = 0). The function “generate_real_samples(n)” randomly selects genuine samples from the dataset, labeling them as “real” (y = 1).
In order to define the structure of the generator model, the function “define_generator(latent_dim, n_outputs)” is utilized. This function employs the Keras sequential model API and consists of two dense layers with the ‘relu’ activation function. The first hidden layer comprised of 15 nodes, while the second hidden layer had 30 nodes. The generator takes latent points as input and produces synthetic samples as output. The sequential model facilitates the creation of a linear stack of layers.
Similarly, the function “define_discriminator(n_inputs)” is used to establish the structure of the discriminator model, also utilizing the Keras library. The discriminator takes input samples, including the counterfeit samples generated by the generator, and evaluates their authenticity. Through its layers, the discriminator extracts features and processes them using weighted connections and activation functions. This transformation enables the capture of relevant information. The discriminator architecture incorporates three hidden layers with the ‘relu’ activation function. The first hidden layer had 25 nodes, the second hidden layer had 50 nodes, and the last layer contained a single node. As the data flows through the discriminator's layers, it gradually learns to differentiate between real and fake samples based on the acquired features. The last layer of the discriminator employs a sigmoid activation function, producing a binary output ranging from 0 to 1. This output represents the probability of the input sample being real or fake, with a value close to 1 indicating high authenticity and a value close to 0 indicating low authenticity. Using multiple layers in the generator and discriminator offers the benefit of enhancing the models' capacity to comprehend and depict intricate patterns within the data. This advantage translates into improved performance, enabling the models to generate more realistic samples and achieve greater accuracy in distinguishing between real and fake samples.38 By increasing the dimension of the nodes in discriminator, it was expected that the network would extract more information from the generator.39
Combining the aforementioned generator and discriminator models results in the construction of the GAN model. The GAN model takes latent points as input, generates counterfeit samples using the generator, and predicts their authenticity using the discriminator. Finally, the program trains the GAN by utilizing a combination of real and counterfeit samples. The discriminator and generator models were alternately trained for 100 epochs. The GAN algorithm was configured to produce an output of three times the size of the data it was generating from. The training progress was monitored through the evaluation of discriminator and generator losses. These losses were visualized in a history plot to provide insights into the dynamics of the GAN model (Fig. A II in ESI†).
To incorporate the three datasets into a unified framework, a low-level data fusion technique was employed. This technique involved stacking the data from different sources on top of each other, resulting in the creation of a new matrix.40 In order to augment the data, two distinct modes were employed, as described in ref. 41:
1. Posterior (post-fusion) to the merging of the datasets (e.g., W + C + DG + GAN)
2. Interstitial (pre-fusion) of the datasets (e.g., W + GAN_W + C + GAN_C + DG + GAN_DG)
Applying GAN to posteriorly merged dataset results in the generation of synthetic data that exhibits variations across different dataset types. On the other hand, the interstitial dataset contains more individual and homogeneous data types.42 Moving forward, the datasets generated through HTL will be referred to as the “original” datasets. The datasets consisting of the original dataset along with the synthetic samples generated by GAN will be referred to as the “hybrid” datasets.
Feature engineering – statistical features
Within the machine learning pipeline, feature engineering plays a pivotal role in enhancing the modeling capabilities of algorithms to effectively fulfil their intended functions. The features were determined by calculating the difference of statistical values at specific wavelengths (represented as “wi”), where each wavelength corresponds to a different statistical value. This technique was partially utilized in the engineering of FTIR spectrum features.28 Furthermore, it was extensively applied in another ML domain, specifically in the field of human activity learning (Table 1).27In general, the application of feature engineering techniques can significantly enhance the accuracy of classifiers for various reasons.
Firstly, these techniques facilitate the capture of crucial distributional properties of the data, assisting classifiers in distinguishing between various classes or patterns. Analyzing the distributional properties of features can also aid in outlier identification and handling. Outliers, being data points that deviate significantly from the majority, have the potential to distort the distribution and impact classifier performance. Detecting and potentially treating or removing outliers can enhance the accuracy of the classification process.43
Secondly, feature engineering techniques can exhibit discriminative power, meaning they possess distinct values for different classes or patterns within the data.44 For example, in the case of spectra of IR spectroscopy, calculating the differences of statistical values can help highlight the unique characteristics of different classes, making it easier for the classifier to differentiate between them.
Thirdly, feature engineering can help reduce the impact of noise, by emphasizing the relative changes in the spectra rather than absolute intensity value.45 For instance, in the context of spectra from IR spectroscopy, the calculation of differences between statistical values can help emphasize the variations that are relevant for classification while reducing the impact of noise or absolute intensity values that may be subject to fluctuations.
Feature extraction via UMAP
The addition of statistical features to a dataset for classification algorithms can also have drawbacks. To begin with, it can lead to the curse of dimensionality, where an excessive number of features hampers algorithm performance, increases complexity, and risks overfitting. Moreover, incorporating statistical features may introduce intricate relationships and interactions among features, making the algorithm harder to interpret. Lastly, irrelevant or redundant features can negatively impact performance and increase the risk of overfitting. Careful consideration, such as feature selection and regularization techniques, is necessary to mitigate these challenges and optimize classification algorithm outcomes.Non-linear feature extraction techniques have demonstrated superior performance compared to classical approaches like linear principal component analysis (PCA) or linear discriminant analysis (LDA) on datasets with a similar tabular structure, such as the time-series ECG200.46 In this study, UMAP method was employed as a feature selection technique to reduce the dimensionality of the dataset, focusing on the most valuable features. Dimension reduction techniques have been found to improve classification performance, prevent overfitting and underfitting of SVC, and enhance the runtime efficiency of the classification algorithm.47 The hyperparameters selection was done by plotting the UMAP results on different purposely selected values for n_neighbors and n_components as it was applied in these studies.48,49 Example of the datapoints distribution is plotted in Fig. A IV in ESI.† A guide to the code used to generate and plot the figure can be found at ref. 50. Following the investigation of hyperparameters, the dimension was embedded with 65 components (n_components) as the default parameter. To ensure a comprehensive overview of the data's overall structure, the size of the local neighborhood (n_neighbors) was limited to 15. This constraint enabled UMAP to effectively capture the inherent structure of the data. Notably, in the context of infrared spectroscopy, the interaction between atoms and infrared radiation occurs across multiple wavenumbers. The Euclidean metric parameter was utilized to control the computed distance between data points.
Data classification
The HPLC results were used to allocate correct labels to the spectrum samples. Samples with a value of 0.6 wt% glucose are considered to have a HTL NER value of 50%. Therefore, when glucose levels were ≥0.6 wt% spectrum samples were labelled as 1, and those below labelled as 0. Several classification algorithms, Adaboost, Gradient Boosting, Random Forest, K (Nearest) Neighbours, SVC and Logistic Regression, were initially scanned to understand the performance across the datasets and their permutations. The classifiers results showed similar performance across the varied datasets (Table AIV in ESI†). However, SVC is commonly employed in model production from infrared datasets. The advantage is given by the projection of input attributes into a high dimensional feature space, thus returning a good generalization when dealing with small datasets.51 Data processing and classification studies were done on laptop machine with an Intel Core i5-6300 CPU 2.40 GHz × 4 processor and 8 GB RAM of memory.Support vector machine classification
The data was split into train and test, 80/20. The data was transformed using the scikit's Min–Max scaler. The hyper-parameters were tuned using the GridSearchCV through the following parameters: regularization parameter, C: 0.1, 1, 10, 100, 1000, tolerance for stopping criterion, tol: 0.005, and rbf kernel. The number of re-shuffling & splitting iterations was set to 10. The metrics used to assess the classification model was average accuracy % over 10 tests.Feature importance
The wavelengths characteristic of solid glucose is shown in the infrared spectrums in Fig. A I(a) in ESI.† With the introduction of water, the characteristic stretching become broader and reduced in intensity (Fig. A I(b), ESI†). In HTL conversion, additional compounds are present in the aqueous phase. These make the spectrums even more challenging to interpret. To determine whether the classification model interprets the data accurately, we need to understand whether the corresponding wavelengths are used to influence the model's accuracy. Therefore, we implement SHAP to visualize which types of features are more or less important. The SHAP library in Python was used to calculate the values using a computer with an AMD Ryzen Threadripper 3906 × 24 – Core Processor 3.79 GHz processor and 128 GB RAM of memory.Results and discussion
Data diversity pre and post GAN
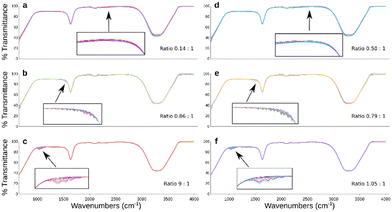
Training only on the underrepresented classes can lead to mode collapse, meaning that the model fails to capture and generate a diverse range of outputs. We confirm that GAN application to each individual dataset resulted in balanced datasets. The ratios of samples above 50% NER to those below tend towards a statistical equilibrium. Visually it is confirmed that the synthetic samples alone follow the profile of the original data (Fig. 2).Classification
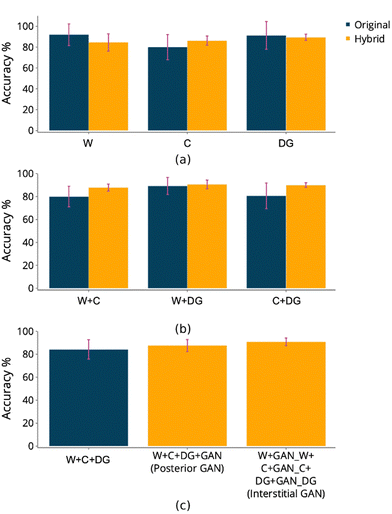
The SVC algorithm was tested against the original and hybrid datasets, individually and then on fused datasets. Wood and dissolved glucose average classification accuracies for hybrids were less than those from the original datasets. Conversely, cotton was slightly higher (Fig. 3(a)). The classification accuracies for fused datasets increased for hybrid compared to the original spectrum samples (Fig. 3(b)). The fusion of the three datasets (W + C + DG) returned a classification accuracy of 84%. This value was lower than the individual original datasets but higher than partial mergers (W + C, W + DG, C + DG). The accuracies for hybrid datasets showed an increase in value. However, it was noticed that the classifier's accuracy depends on the fusion type. A posterior GAN application returns a lower accuracy than the interstitial GAN application, 88% compared to 91%, respectively (Fig. 3(c)). The SVC classifier appears to perform relatively better on the interstitially fused dataset in terms of precision, recall and F1 score. The W + C + DG dataset had lower but still decent performance, while the W + C + DG + GAN dataset showed the lowest performance across all metrics (Table 2). Additional details of the initial study are highlighted in Table V and confusion matrices, shown in Fig. A III in ESI.†| Dataset | Precision | Recall | F1 score | Accuracy/% |
|---|---|---|---|---|
| W + C + DG | 0.9397 | 0.7790 | 0.8382 | 84 |
| W + C + DG + GAN | 0.7594 | 0.3429 | 0.4696 | 88 |
| W + GAN + C + GAN + DG + GAN | 0.9286 | 0.8592 | 0.8903 | 91 |
Ablation study
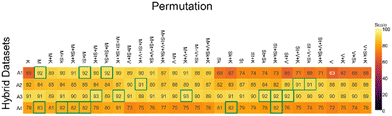
It has been reported that feature engineering can further improve the accuracy of the classifiers. Different dataset permutations encompassing the additional statistical features were explored. Additionally, UMAP was applied to reduce the effect of low contributing values such as those derived from the water content and other compounds, and the accuracies reported.The accuracy of the base model 1, which is the hybrid dataset with GAN applied posteriorly to the merging (W + C + DG + GAN) was 88%. In the first ablation study (ablation study A1, Fig. 4), fifteen permutations showed more than 10% decrease in accuracy, two returned similar values, while the others manifested incremental increases, with three reaching 92% (highlighted by green borders). For the most performing models, this represents a 4% increase in accuracy compared to the base model 1. UMAP application (ablation study A2, Fig. 4) showed similar performance to the base model 1, with the exception of three outliers that reached 91% accuracy. Interestingly, UMAP stabilized the performance of those models that were fitting poorly earlier in ablation study A1. This could be the result of retaining only the glucose contributory features.
The accuracy of the base model 2, which is the hybrid dataset with GAN applied interstitially (W + GAN_W + C + GAN_C + DG + GAN_DG) was approximately 91%. The process of feature engineering improves the models to 92% and 93% respectively, a small but valuable contribution to the classification of glucose in aqueous solution (ablation study A3, Fig. 4). UMAP application (ablation study A4, Fig. 4) performed poorly compared to base model 2, reducing the accuracies to 70 to 80% range.
Model selection
The current framework showed increased accuracy results compared to the model trained on the original dataset only (W + C + DG). At the same time, the study showed that numerous models could be used for the portrayed application. Therefore, further refinement for the best model was conducted and it involved a selection based on additional performance metrics such as precision and recall. Some of the most performant datasets were picked using the accuracies reported in the ablation study (Fig. 4). We calculated the average performance metrics over ten iterations (some are shown in Table 3 and additional ones Table VI in ESI†). The selection criteria aimed at finding a model with balanced precision and recall, which would make it suitable for classifying samples with hardly distinguishable compounds. Based on this criterion, the W + GAN_W + C + GAN_C + DG + GAN_DG + M + V + K dataset was the most suitable model for our application.| Dataset permutation | Accuracy/% | Standard deviation | Precision | Recall | |
|---|---|---|---|---|---|
| A1 | M | 91.80 | 2.88 | 0.9737 | 0.8981 |
| A2 | St + V + Sk | 90.96 | 3.64 | 0.9237 | 0.9254 |
| A3 | M + V + K | 93.49 | 2.35 | 0.9588 | 0.9313 |
| A4 | M + Sk | 81.56 | 4.50 | 0.8873 | 0.8070 |
Model explainability
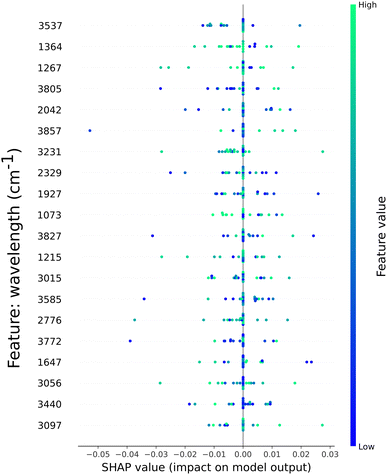
In this section, we present the feature importance as calculated by SHAP for two classification models. We visualized the top 20 most important features, as sorted by SHAP, out of 900 available. To highlight the differences between a non-processed dataset and post processed, we selected two models and plotted the SHAP values for each.In Fig. 5 the SHAP values and their contribution to the classification model based on the W + C + DG dataset are shown. Absorption values from across the spectrum are present, from the O–H group characteristic to 3000 to 4000 cm−1 (10 out of 20 features) stretching and C–O group stretching. The impact of these features is shown by the coloured dots. Preponderant high values are present in the absorption of the C–H stretching in CH3 at ∼1364 cm−1, the syringyl ring breathing represented at ∼1267 cm−1 and ∼1215 cm−1, as well as the C–O stretching and ∼1073 cm−1, respectively. The O–H group stretching has lesser impact towards the model output, as highlighted by the blue dots. The even distribution of features impact might be the reason for the average accuracy of 84%.
In Fig. 6, the feature contribution of the dataset W + GAN_W + C + GAN_C + DG + GAN_DG + M + V + K (highlighted in Table 3) are presented. The classification model using this dataset showed the highest performance accuracy, an average of 93.49% ± 2.35%.
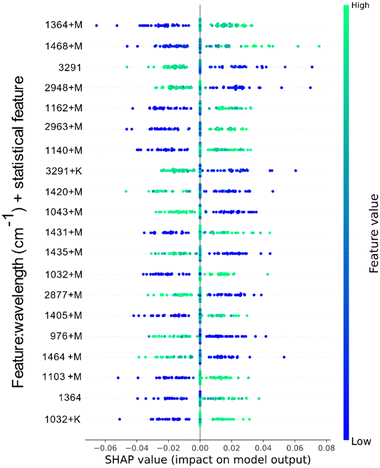 | ||
| Fig. 6 Top 20 features contribution towards the model classification for the W + GAN_W + C + GAN_C + DG + GAN_DG + M + V + K dataset. | ||
Compared to the model in Fig. 5, feature engineering played a more significant role in the order of importance of values. In this case, only two values from the original dataset are part of the top 20 most important features, namely the absorption from the O–H stretching, ∼3291 cm−1, and the aliphatic C–H stretching in CH3, ∼1364 cm−1. The primer has negative impact on the classification model, whereas the secondary has a high positive impact. Having the C–H stretching contributing towards the model is valuable, since this stretching is part of the glucose composition as seen in Fig. A I (in ESI†). The resulted mean difference feature from the same wavelength is also positively influencing the model output, and it tops as the most influential feature (∼1364 cm−1 + M). Similarly, the engineered feature of the O–H stretching containing the kurtosis difference value is also showing a negative contribution towards the model output. Other significant features captured in the top 20 most important features contributing to the model output include the mean difference of compounds in the frequencies ∼1468 cm−1, ∼1162 cm−1, ∼2963 cm−1, ∼1140 cm−1, ∼1431 cm−1, ∼1032 cm−1, ∼1405 cm−1, 1103 cm−1, and kurtosis difference of 1032 cm−1. These frequencies directly express those seen in solid and aqueous glucose solution spectrums. Additionally, it can be noticed that O–H stretching range and C–O group range (3000 to 4000 cm−1) contribute negatively to the model output. This is because they represent groups of compounds found in water, which are not important in identifying compounds specific to glucose.
Conclusions
This study investigated the use of infrared spectrums as data support for classifying glucose production within an HTL conversion system, a green technology for generating renewable energy. As a result, a novel method for improving the model accuracy using synthetic data generation was developed. Initially, a low number of test runs using wood and cotton were produced using HTL. High-purity glucose was also dissolved in deionized water to increase the dataset, in addition to the laborious HTL output.First, individual datasets were used for building a classification model. Second, GAN was applied under two data fusion circumstances. It was found that the classification of the hybrid datasets is dependent on the fusion type. GAN used posteriorly scored a lower accuracy compared to GAN applied interstitially. Furthermore, hand-crafted features were added to improve the classification models. The results showed an average accuracy increase of more than 9% over the base model, from 84% to more than 93%. Under the same argument, we also applied UMAP. The dimensionality reduction method did not exceed the earlier reported accuracy but it improved above the base model from 84% to 91%. The best performing model was explained by employing SHAP values. It was found that within the top 20 features, those related to the glucose compounds are positively influencing the classification model, whereas those found in water are negatively contributing towards the model output. Although this framework is tested on the HTL biomass conversion system, it opens new avenues for integrating FTIR in continuous process monitoring.
For example, the integration of data augmentation using generative AI and IR spectroscopy for process monitoring has the potential to revolutionize costly and lengthy research and development activities such as monoclonal antibody production, gene therapy manufacturing, and cultured meat production. Generative AI techniques enable the generation of synthetic data, augmenting existing datasets and providing greater volume and variability. This augmented dataset improves machine learning model training, enhancing accuracy and generalization. Consequently, it accelerates the research cycle by enabling simulation, prediction, and optimization of process parameters without extensive physical experimentation. FTIR as a sensory technique allows real-time process monitoring, continuously analyzing critical quality attributes and parameters to ensure consistency, reproducibility, and early detection of deviations. When coupled with a classifier such as SVC, can even outperform traditional process control techniques (e.g., Proportional–Integral–Derivative). This enables timely interventions and corrective actions, reducing batch rejections and enhancing overall product quality. Ultimately, the implementation of generative AI and IR spectroscopy mitigates risks in the aforementioned research and development activities, resulting in cost savings by minimizing production failures and optimizing process performance.
Consequently, the current method offers the distinct benefit of being a decentralized AI system, addressing the issue of biases found in master datasets. Master datasets, typically sourced from large-scale platforms, may unknowingly harbor biases and dominant features that contribute to inequalities or reinforce societal imbalances. However, by training decentralized AI models using local data, such as the data generated under the HTL conditions outlined in this study, this method potentially mitigates these biases and fosters fairer and more inclusive machine learning applications.
Author contributions
Silviu Florin Acaru: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft. Rosnah Abdullah: funding acquisition, project administration, supervision. Daphne Teck Ching Lai: resources, supervision, writing – review & editing. Ren Chong Lim: project administration, supervision, writing – review & editing.Conflicts of interest
There authors have no competing interests.Acknowledgements
The authors would like to thank Parham Hadikhani for helpful discussions related to deploying deep learning techniques. The work carried out in this article was supported using a University Research Grant (URG) from Universiti Brunei Darussalam (UBD/RSCH/URC/RG(b)/2019/013). Silviu Florin Acaru is a recipient of the UBD University Graduate Scholarship (UGS).Notes and references
- X. Zhuang, J. Liu, C. Wang, Q. Zhang and L. Ma, Fuel, 2022, 317, 123462, DOI:10.1016/j.fuel.2022.123462.
- A. Taghipour, U. Hornung, J. A. Ramirez, R. J. Brown and T. J. Rainey, J. Cleaner Prod., 2021, 289, 125582, DOI:10.1016/j.jclepro.2020.125582.
- X. Gu, N. Pang, Y. Qiu, X. Fu, Y. Yao and S. Chen, Fuel, 2022, 310, 122358, DOI:10.1016/j.fuel.2021.122358.
- H. Aljabri, P. Das, S. Khan, M. AbdulQuadir, M. Thaher, A. H. Hawari and N. M. Al-Shamary, Renewable Energy, 2022, 189, 78–89, DOI:10.1016/j.renene.2022.02.100.
- X. Ding, S. Mahadevan Subramanya, K. E. Waltz, Y. Wang and P. E. Savage, Bioresour. Technol., 2022, 352, 127100, DOI:10.1016/j.biortech.2022.127100.
- C. Hong, Z. Wang, Y. Si, Z. Li, Y. Xing, J. Hu and Y. Li, Sci. Total Environ., 2021, 776, 145596, DOI:10.1016/j.scitotenv.2021.145596.
- H. Wang, Y. Jiang, E. Park, X. Han, Y. Zeng and C. Xu, Sustainability, 2023, 15(8), 6698, DOI:10.3390/su15086698.
- M. El Bast, N. Allam, Y. Abou Msallem, S. Awad and K. Loubar, J. Energy Inst., 2023, 108, DOI:10.1016/j.joei.2023.101260.
- S. F. Acaru, R. Abdullah, D. T. C. Lai and R. C. Lim, Heliyon, 2022, 8, e10738, DOI:10.1016/j.heliyon.2022.e10738.
- S. F. Acaru, R. Abdullah and R. C. Lim, Waste Biomass Valorization, 2023 DOI:10.1007/s12649-023-02074-y.
- P. S. Fomina, M. A. Proskurnin, B. Mizaikoff and D. S. Volkov, Crit. Rev. Anal. Chem., 2022, 1–18, DOI:10.1080/10408347.2022.2041390.
- B. K. Mekonnen, W. Yang, T. H. Hsieh, S. K. Liaw and F. L. Yang, Biomed. Signal Process. Control, 2020, 59, 101923, DOI:10.1016/j.bspc.2020.101923.
- Y.-T. Wang, B. Li, X.-J. Xu, H.-B. Ren, J.-Y. Yin, H. Zhu and Y.-H. Zhang, Food Chem., 2020, 303, 125404, DOI:10.1016/j.foodchem.2019.125404.
- E. Korb, M. Bağcıoğlu, E. Garner-Spitzer, U. Wiedermann, M. Ehling-Schulz and I. Schabussova, Biomolecules, 2020, 10, 1–17, DOI:10.3390/biom10071058.
- D. M. Mackie, J. P. Jahnke, M. S. Benyamin and J. J. Sumner, MethodsX, 2016, 3, 128–138, DOI:10.1016/j.mex.2016.02.002.
- D. L. Sills and J. M. Gossett, Biotechnol. Bioeng., 2012, 109, 353–362, DOI:10.1002/bit.23314.
- J. Chen, L. Wu, T. Pan, J. Xie and H. Chen, in 2010 Seventh International Conference on Fuzzy Systems and Knowledge Discovery, IEEE, 2010, pp. 2159–2163 DOI:10.1109/FSKD.2010.5569754.
- R. B. Madsen, K. Anastasakis, P. Biller and M. Glasius, Energy Fuels, 2018, 32, 7660–7669, DOI:10.1021/acs.energyfuels.8b01208.
- Y. Xu, J. Ge and C.-W. Ju, Energy Adv., 2023 10.1039/D3YA00057E.
- C. Sui, Y. Y. Li, X. Li, G. Higueros, K. Wang, W. Xie and P. C. Hsu, Adv. Energy Mater., 2022, 12(6) DOI:10.1002/aenm.202103044.
- Z. Ullah, S. Raza, W. Farooq, H. Yang, S. Wang and D. N. Vo, Bioresour. Technol., 2021, 335, 125292, DOI:10.1016/j.biortech.2021.125292.
- T. Katongtung, T. Onsree and N. Tippayawong, Bioresour. Technol., 2022, 344, 126278, DOI:10.1016/j.biortech.2021.126278.
- S. Motamed, P. Rogalla and F. Khalvati, Inform. Med. Unlocked, 2021, 27, 100779, DOI:10.1016/j.imu.2021.100779.
- B. Bosquet, D. Cores, L. Seidenari, V. M. Brea, M. Mucientes and A. Del Bimbo, Pattern Recognit., 2023, 133, 108998, DOI:10.1016/j.patcog.2022.108998.
- Q. Zhu, B. Sun, Y. Zhou, W. Sun and J. Xiang, IEEE Trans. Instrum. Meas., 2021, 70, 1–10, DOI:10.1109/TIM.2021.3077995.
- X. Ma, K. Wang, K. C. Chou, Q. Li and X. Lu, Anal. Chem., 2022, 94, 577–582, DOI:10.1021/acs.analchem.1c04263.
- D. A. Adama, A. Lotfi, C. Langensiepen, K. Lee and P. Trindade, Soft comput., 2018, 22, 7027–7039, DOI:10.1007/s00500-018-3364-x.
- S. Toraman, M. Girgin, B. Üstündağ and İ. Türkoğlu, Turk. J. Electr. Eng. Comput. Sci., 2019, 27, 1765–1779, DOI:10.3906/elk-1801-259.
- L. McInnes, J. Healy and J. Melville, arxiv, 2018, preprint, arXiv:1802.03426 DOI:10.48550/arXiv.1802.03426.
- S. M. Lundberg and S.-I. Lee, in Advances in Neural Information Processing Systems, ed. I. Guyon, U. Von Luxburg, S. Bengio, H. Wallach, R. Fergus, S. Vishwanathan and R. Garnett, Curran Associates, Inc., 2017, vol. 30 Search PubMed.
- A. S. Rathore, S. Nikita, G. Thakur and N. Deore, Curr. Opin. Chem. Eng., 2021, 31, 100671, DOI:10.1016/j.coche.2021.100671.
- H. Tiernan, B. Byrne and S. G. Kazarian, Spectrochim. Acta, Part A, 2020, 241, 118636, DOI:10.1016/j.saa.2020.118636.
- I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville and Y. Bengio, Commun. ACM, 2020, 63, 139–144, DOI:10.1145/3422622.
- M. Arjovsky, S. Chintala and L. Bottou, 34th International Conference on Machine Learning, ICML 2017, 1, 298–321.
- I. Gulrajani, F. Ahmed, M. Arjovsky, V. Dumoulin and A. Courville, Adv. Neural Inf. Process. Syst., 2017, 5768–5778 Search PubMed.
- L. Xu and K. Veeramachaneni, arXiv, 2018, preprint, arXiv:1811.11264 DOI:10.48550/arXiv.1811.11264.
- N. Park, M. Mohammadi, K. Gorde, S. Jajodia, H. Park and Y. Kim, Proceedings of the VLDB Endowment, 2018, vol. 11, pp. 1071–1083 DOI:10.48550/arXiv.1806.03384.
- S. F. Ahmed, M. S. Bin Alam, M. Hassan, M. R. Rozbu, T. Ishtiak, N. Rafa, M. Mofijur, A. B. M. Shawkat Ali and A. H. Gandomi, Artif. Intell. Rev., 2023 DOI:10.1007/s10462-023-10466-8.
- D. Stathakis, Int. J. Remote Sens., 2009, 30, 2133–2147, DOI:10.1080/01431160802549278.
- T. Casian, B. Nagy, B. Kovács, D. L. Galata, E. Hirsch and A. Farkas, Molecules, 2022, 27, 4846, DOI:10.3390/molecules27154846.
- Q. Liu, G. Ma and C. Cheng, IEEE Access, 2020, 8, 70111–70124, DOI:10.1109/ACCESS.2020.2986356.
- P. Liu, J. Li, L. Wang and G. He, IEEE Geosci. Remote Sens. Mag., 2022, 10, 295–328 Search PubMed.
- V. Rizeakos, A. Bachoumis, N. Andriopoulos, M. Birbas and A. Birbas, Appl. Energy, 2023 DOI:10.1016/j.apenergy.2023.120932.
- E. S. Sabry, S. S. Elagooz, F. E. A. El-Samie, N. A. El-Bahnasawy, G. M. El-Banby and R. A. Ramadan, J. Opt., 2023, 52, 716–741, DOI:10.1007/s12596-022-01024-6.
- S. Zhang, P. Rao, H. Zhang, X. Chen and T. Hu, Infrared Phys. Technol., 2023, 132 DOI:10.1016/j.infrared.2023.104670.
- F. Anowar, S. Sadaoui and B. Selim, Comput. Sci. Rev., 2021, 40, 100378, DOI:10.1016/j.cosrev.2021.100378.
- UMAP as a Feature Extraction Technique for Classification—umap 0.5 documentation, https://umap-learn.readthedocs.io/en/latest/auto_examples/plot_feature_extraction_classification.html, (accessed 26 November 2022).
- M. Cihan Sorkun, D. Mullaj, J. M. V. A. Koelman and S. Er, Chem.: Methods, 2022, 2(7) DOI:10.1002/cmtd.202200005.
- S. Bej, J. Sarkar, S. Biswas, P. Mitra, P. Chakrabarti and O. Wolkenhauer, Nutr. Diabetes, 2022, 12, 1–11, DOI:10.1038/s41387-022-00206-2.
- Basic UMAP Parameters—umap 0.5 documentation, https://umap-learn.readthedocs.io/en/latest/parameters.html, (accessed 26 November 2022).
- O. Devos, C. Ruckebusch, A. Durand, L. Duponchel and J. P. Huvenne, Chemom. Intell. Lab. Syst., 2009, 96, 27–33, DOI:10.1016/j.chemolab.2008.11.005.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ya00236e |
| This journal is © The Royal Society of Chemistry 2023 |