Open Access Article
Open Access ArticleRoute selection and reaction engineering for sustainable metabolite synthesis†
Roland
Wohlgemuth
 *abc
*abc
aMITR, Institute of Applied Radiation Chemistry, Faculty of Chemistry, Lodz University of Technology, 90-537 Lodz, Poland. E-mail: roland.wohlgemuth.1@p.lodz.pl
bSKB, Swiss Coordination Committee Biotechnology, 8021 Zurich, Switzerland
cESAB, European Society of Applied Biocatalysis, Brussels, Belgium
First published on 30th June 2023
Abstract
The great advances in information technology, machine learning and artificial intelligence are providing fast access to potential routes to a desired metabolite from possible starting materials. While routes can be easily ranked according to clearly measurable indicators, such as the number of reaction steps involved, reaction chemistry-based knowledge and experience of what is working and also what is not working is important as well. Multiple factors such as availability and sources of starting materials, reaction selectivity, safety, resource-efficiency and economy, influence the selection of a route to be developed. Sustainability aspects of synthetic methodologies have also become additional selection criteria for routes of biological and chemical reactions to metabolites. Analytical methodologies are thereby essential not only for rapid prototyping but also for analyzing the operational windows for the reactions of the prototype route and for finding optimum reaction conditions, which can themselves be used for further reaction optimization in an iterative way.
Introduction
The small molecular weight compounds of living cells represent, in addition to the DNA, RNA and protein space, an important molecular space of biology attracting increased attention with the renewed interest in cellular metabolism and its regulation in health and disease.1,2As investigations of the entire metabolite space have in this context come into focus, gaps have become clear between how many metabolites have been identified in a biological organism and how many of these have already been described to be synthesized in the laboratory. Despite numerous past chemistry and biochemistry milestones which have been achieved in connection with obtaining desired metabolites, many essential metabolites are not available, and therefore metabolite synthesis experienced a renaissance as access to pure metabolites is a key requirement.3 Metabolites are needed for life to exist, as clearly evident from the dependence of cellular growth on metabolites as nutrients in defined culture media and the environment. Even for the newest field of synthetic biology metabolites continue to be key to the growth of organisms that have been repurposed with synthetic genomes. The impressive work on synthetic genomes, which were transplanted into a favourable cellular environment for bringing them to life, depended also on the supply in the medium of practically all the required metabolites.4,5 The discovery, analysis and identification of easily diffusable metabolites, which show a large diversity of structures and biological functions, have become major tasks. This has lead to important advances in understanding cellular metabolism, a significant growth of the number of known metabolite structures which have so far been identified in various biological organisms, valuable tools for their contextual mapping and visualization, as well as the development of better data storage and information systems.6–10 As interactions of a specific biological organism with its associated microbial cells are of importance for the biological organism's health status and involve also bioactive small molecules, it is of much interest not only to describe the metabolome of the biological host organism but also to start cataloguing the metabolites of its corresponding microbiome, for example in case of the Human Metabolome Database HMDB9 the corresponding Human Microbial Metabolome Database MiMeDB.10 The rapidly growing number of identified metabolites and the much slower growth in the number of the synthesized metabolites are illustrated by the example of the human metabolite database HMDB.9 In line with these developments, the synthesis of metabolites has therefore also moved forward very much. While many metabolites have been manufactured at large scale for various applications, smaller scales are adequate for numerous other natural and non-natural metabolites. While suitable inhouse facilities for these projects are of interest to large organisations, it is worth mentioning that there are options of having analytical or synthetic work on metabolites performed by specialized service companies, for example providing a service for metabolite synthesis (for a selection of service institutions see Table S1 in the ESI†), or for the analysis and identification of metabolites (for a selection of service institutions see Table S2 in the ESI†).
The continuous increase in the number and scale of metabolite synthesis over the years has not only brought more attention to the optimization of space–time yields, but also to the availability of less abundant biological or elemental resources, resource efficiency to manufacture more with less, safety, health and various environmental aspects. The concept of sustainable development, described in the Gro Harlem Brundtland report more than 40 years ago,11 and the straightforward description of the mass of waste per mass of product synthesized with the E factor, introduced by Roger Sheldon more than 30 years ago,12 have been crucial for guiding developments and technologies in general and in particular also for considering the sustainability of metabolite manufacturing processes. Whether metabolites are isolated from biological raw materials, produced by fermentation or by chemical synthesis the molecular economy strategies of resource-efficient processes preserving biodiversity and limited elements and the prevention of waste accumulation have contributed to greener processes in the synthesis of metabolites. Considering the transition from fossil-based raw materials to bio-based resources as a key direction, and in view of the numerous renewable bioprocesses in the biosphere, implementing bioeconomy strategies for utilizing biobased resources and bioprocesses is also important for the sustainable development of metabolite synthesis, as metabolites are manufactured in various industrial sectors.13 As biobased resources differ from fossil-based raw materials by the presence of many functional groups, highly selective biocatalytic defunctionalization reactions14 can provide a much shorter synthetic route to valuable metabolites than the introduction of new functional groups into highly reduced fossil-based starting materials.
Over the whole workflow (see Fig. 1), from the design phase over rapid prototyping, reaction engineering and product recovery and purification to scaling, advanced strategies in biocatalysis, metabolic engineering and synthetic biology have contributed to better production processes in metabolite synthesis. These are not only more sustainable than less resource-efficient and longer chemical routes, which in addition may have safety, health and environmental issues, but also have sustainability benefits in comparison with biological routes to metabolites formed in only small and varying amounts naturally in biological species and thus also having an unfavorable E factor. Therefore it is important to consider the concept of safe and sustainable by design.15
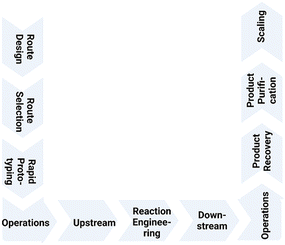 | ||
| Fig. 1 Workflow for metabolite synthesis from route design and selection, rapid prototyping, reaction engineering, upstream and downstream operations, product recovery and purification to scaling. | ||
Designing and selecting a sustainable synthesis route to a desired metabolite target is best done at the beginning before scaling. Benefits may however also be achieved by redesign of the process in the course of the transfer from laboratory scale to production, or by improving or replacing a manufacturing process which is already established at industrial large scale. This is illustrated by the changes from chemical processes to bioprocesses in the manufacturing of vitamin B12 (ref. 16) and vitamin B2 (ref. 17) at industrial large scale, where sustainability continues to be a driver for process improvements. A more sustainable route provides also added benefits such as improvements in safety and health, efficiency of human, energy and material resources. This translates into continuing economic benefits, which can counterbalance the one-time effort of changing within the same company an industrial process even at large scale. Alternatively, this may provide an opportunity for a competitor to develop an improved manufacturing process to large scale.
Reaction engineering, upstream and downstream processing are essential elements in the development of a viable metabolite manufacturing process via the selected route.
Design and selection of target-oriented synthesis routes
The experimental knowledge base of metabolite synthesis has continuously grown over time, from Friedrich Wöhler's mile-stone discovery that the organic metabolite urea, which is naturally formed in kidneys of humans and animals, can be synthesized in the laboratory from inorganic ammonium chloride and silver cyanate,18 to the synthesis of numerous organic compounds and bioactive small molecules occurring naturally in biological organisms. Impressive total syntheses of natural products with increasing molecular complexity and diversity have been achieved by tremendous advances in the science and art of target-oriented synthesis.19 Metabolite synthesis represents a hallmark of organic chemistry and many new synthetic methodologies have been developed in the course of finding a total synthesis route to a desired metabolite/natural product target.20–22The access to a tremendous experimental knowledge about which synthetic reactions have worked well in the past, but also which synthetic transformations did not work well or failed, continues to be an important prerequisite for synthesis planning. This has been pioneered with the concept of the retrosynthetic analysis and its disconnection of bonds by E. J. Corey,23 by brainstorming and creatively thinking backwards from a desired target compound via a single reaction step in terms of chemical reactions to a precursor from which it could be made. This iterative approach until a commercially available starting compound is reached has been envisioned very early to benefit from combining chemical retrosynthetic analysis with information technology, combining the use of the strategies of retrosynthetic analysis and an adequate database in a computer program.24
Computer-assisted synthesis planning has progressed very much since then, and advanced software tools using data-driven models25 and rule-, similarity-, or transformer-based models.26–28 Based on large digital chemical reaction databases a number of commercial solutions to the problem are offered, such as the CAS SciFindern – Retrosynthesis software,29 the Reaxys Predictive Retrosynthesis software,30 and the Chematica/SYNTHIA™ Retrosynthesis software.27 An analogous backward thinking approach can also be applied to stereoselective biocatalytic reactions, based on experimentally established enzymatic reaction platforms,31 and the interfacing of biocatalysis and organic chemistry in retrosynthetic analysis has attracted much interest.32–36 Taking a step back to a general level, the visualization of the encyclopedia of known metabolic pathways from nature, the expansion of biochemical knowledge and synthetic metabolic pathways provide another great opportunity for retrosynthetic analysis in the biochemical domain, which has also been designated as retrobiosynthesis.37–40
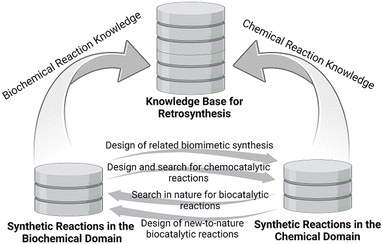
The interactions between the biochemical and chemical domains have been mutually beneficial for the design of target-oriented synthesis routes (see Fig. 2), such as the inspirations from the biochemical pathways to the design of related biomimetic syntheses in organic chemistry,41–43 the inspirations from classical reactions of organic chemistry to the search or design of corresponding natural or new-to-nature biocatalytic reactions or reaction modules,44,45 or the anticipation of a potential natural product and its biosynthesis from its chemical synthesis in the laboratory.46
Although retrosynthetic analysis in the biochemical and chemical domains has been a creative process driven by human experience, existing knowledge and intuition with the support of information technology and machine learning, combining these in a hybrid search is of much interest.47 The consideration of all biochemical and chemical synthetic methods provides a broad and complementary reaction space for search strategies towards suitable and sustainable routes. As a first step in the way forward to a computer-assisted workflow for synthesis planning which incorporates enzymatic as well as organic chemistry based transformations, a single-step retrosynthesis strategy has been demonstrated by the use of natural enzymatic transformations.48
Rapid prototyping of product synthesis
Global supply chain issues, critical product needs related to multiple crises and the emerging bioeconomy49 bring the attention to the development of methodologies and tools for rapid responses in order to introduce more resilience into product synthesis. The robustness of metabolism of biological cells for the synthesis of metabolites at the right place and time when needed can thereby be a blueprint and inspiration for developing a sustainable industrial metabolism. As both biological cells and industrial organisations interact with the environment, some analogy and important common aspects can be fruitful for developments towards a sustainable industrial metabolism. Interconnecting raw material availability, handling of waste, cycles of elements and their regulation, are not only relevant for cellular metabolism but are also essential for activities of industrial organisations. The symbiotic cooperation of different biological cells in a consortium has its counterpart in the cooperation within and across industrial sectors and the attention to resource-efficient material and energy flows in value chains, from raw materials to intermediates and final products. Sustainable synthetic methodologies are thereby not only of much interest for developing and manufacturing natural metabolites, but also metabolite-like non-natural pharmaceuticals in case of critical defense needs against emerging human diseases.Rapid prototyping as a rapid response methodology can also be valuable in product synthesis. The realization of any major bottlenecks at an early development stage can avoid time-consuming detours, and the implementation of the selected synthesis route to the target product at small scale can give important insight into further research needs.50 Impressive advances in developing whole cell systems for metabolite synthesis have been achieved by key discoveries, methodologies and achievements in metabolic engineering over the last three decades51 and by synthetic biology technologies over the last two decades.52 First prototype pathways for demonstrating the synthesis of the desired metabolite in whole cells can thereby be developed faster, and subsequent improvements in yields and product concentrations can be performed by the iterative application of design–build–test–learn (DBTL) cycles.53–55 Key for engineering cellular metabolism towards successful rapid prototyping and increased productivity rather than towards increasing complexity is thereby a sufficiently detailed understanding of the selected whole cell biosystem at molecular, cellular and process levels and the various factors influencing rates, titers and yields.56 Microbial strain design and optimization for metabolite production can be accelerated by new tools integrating computational design, machine learning and manual curation of the knowledge base by humans, as shown for hundreds of metabolites by using the Lila system and Escherichia coli and Saccharomyces cerevisiae as hosts.57 A decisive prerequisite is the availability or development of meaningful and sensitive analytical methods with adequate information content.
Databases containing experimentally measured features and influencing factors such as enzyme functions, kinetics and thermodynamics of enzyme-catalyzed reactions, interactions of substrates and products with enzymes and whole cells, are key for successful DBTL cycles and a prerequisite for using machine learning techniques.58
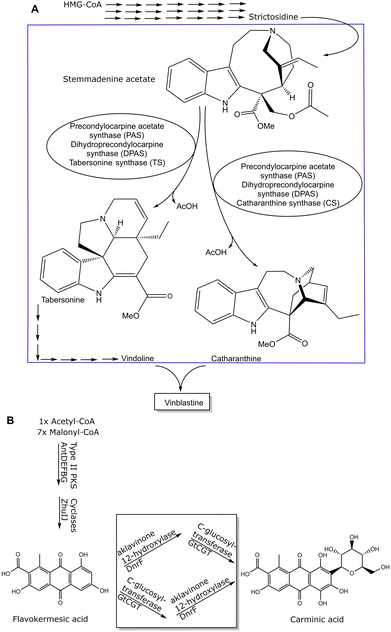
Whole cell approaches for rapid prototyping of metabolite synthesis may have to overcome stability or transport limitations, or one or more enzymes of the pathway to the product are unknown. Therefore, a systems biocatalysis approach provides a combined top-down and bottom-up framework which is useful for investigating a modular set up of reactions, including biochemical and chemical reactions, for achieving sustainable syntheses of metabolites.59,60 It is highly desirable to find more sustainable routes based on the pathways from nature as prototypes in cases when metabolites can only be isolated in limited amounts from natural resources and are thereby creating much waste. As this requires however the knowledge of all enzymes involved in the respective pathway, the identification and characterization of any missing enzyme of that biosynthetic pathway is absolutely essential. Great advances have been achieved in identifying missing enzymes in biosynthetic pathways, as illustrated in Fig. 3 for missing enzymes (see box) in the biosynthetic pathway to vinblastine61 and carminic acid,62 which can serve as blueprints for rapid prototyping.
Cell-free biocatalysis is of much interest for rapidly prototyping metabolite synthesis systems with reduced complexity63–65 compared to whole cell systems. The use of whole cells can however be a great advantage in the reaction engineering to enhanced conversion and yield over cell-free systems, when enzyme stability or reactant replenishment is higher in vivo. Designing and applying cell-free biocatalysis for metabolite production is attractive not only because of simplified downstream processing compared to metabolite purification from the fermentation broth, but also because of the absence of transport boundaries of cell membranes for substrates and products. Potential stability issues of metabolite production using recombinant whole-cell systems may be resolved in a more modular way when selecting cell-free systems for prototyping.
The great progress in DNA synthesis66 and cell-free protein synthesis,67–69 and their use for the prototyping of enzymatic reactions,70–72 are additional key factors for making a rapid design of in vitro biocatalytic systems attractive. A miniaturized workflow using in vitro transcription/translation and an enzymatic assay at microliter scale enabled the rapid prototyping of O-methyltransferase-catalyzed methylation73 towards the substrates caffeic acid, catechol, dopamine, quercetin and ferulic acid. Numerous sustainable syntheses of metabolites have been prototyped by a single enzymatic reaction step (see Fig. 4) from suitably selected and easily available starting compounds using isolated recombinant enzymes.74–80
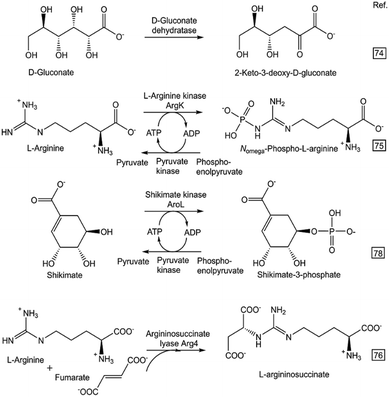 | ||
| Fig. 4 Rapid prototyping of selected one-step enzymatic syntheses of metabolites (described in more details in the indicated literature references). | ||
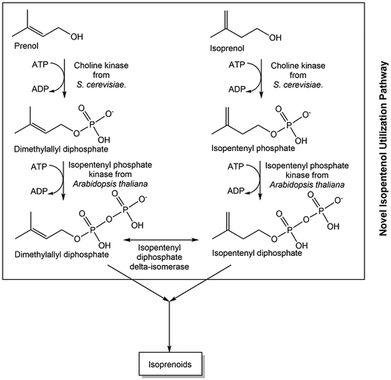
For diverse metabolites with very long and complex biosynthetic pathways, the reduction of complexity by designing a novel shorter pathway module for a common part of the natural pathways is very promising, such as the starting point for the isopentenyl utilization pathway module (see Fig. 5) for isoprenoids.81
Impressive achievements have also been obtained by a combination of enzymatic reactions to a metabolite and its further chemical conversion to the desired metabolite,82 or the chemical coupling of the two metabolites catharanthine and vindoline (see Fig. 3A), which have both been enzymatically synthesized in yeast, to the naturally occurring monoterpene indole alkaloid vinblastine by using Fe(III)-based or photo-chemical coupling methods.83
The advantages of performing multiple reaction steps in one pot without the isolation and purification of intermediates have attracted tremendous interest due to the excellent selectivity and interoperability of suitable isolated recombinant enzymes. This has been demonstrated very early by the 17-step enzymatic synthesis of hydrogenobyrinic acid from 5-amino-levulinic acid in 20% overall yield using 12 recombinantly expressed enzymes of vitamin B12 biosynthesis.84 The reduced complexity and the modular setup of a multistep enzymatic synthesis enables a step by step approach combining experiments and modelling for optimizing the overall conversion, taking into account proper balancing of enzyme activities and avoiding enzyme inhibition.85 Enzymatic reaction sequences have been assembled in a practical and simple approach for rapid prototyping and optimization in vitro by using cell lysates enriched with enzymes which have been prepared by cell-free protein expression.86 The use of cell-free biocatalysis in multistep enzymatic synthesis of metabolites and natural products is not only of interest for the early development phase, but also for reaction engineering and the translation to sustainable manufacturing processes.87,88
Reaction engineering
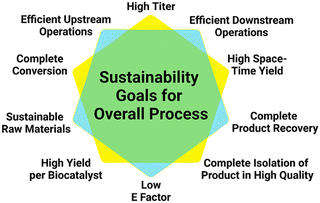
The successful demonstration of a prototype reaction for the product synthesis becomes the starting point for reaction engineering. Significant challenges are thereby the expanding number of options for selecting the optimum settings for operating the reaction and rules for guiding the selection of the optimum path.89 The enzyme-based reactions may be optimized in a whole-cell system, a cell-free system, or a combination thereof, depending on the comparison of the bioproduction systems for a particular metabolite.90 Platform technologies for enzyme-catalyzed reactions, such as enzyme engineering, metabolic engineering, synthetic biology, or immobilization are important for optimizing enzyme-based reactions in whole-cell systems as well as cell-free systems.91 Impressive improvements of titers have been achieved in biocatalytic plant metabolite synthesis by engineering and optimizing their pathways in S. cerevisiae whole cells, for example in the synthesis of the vinblastine precursor metabolites catharanthine and vindoline.17 Some important goals of reaction engineering for metabolite syntheses are highlighted in Fig. 6 and include a high degree of substrate conversion, a high isolated yield of the final metabolite, increasing substrate concentrations for achieving high product space–time yields, high product concentrations, a high final amount of metabolite per amount of biocatalyst and a reduced E-factor,92 while keeping the high product quality which has been achieved in the prototype reaction.The systematic reaction optimization by design of experiments provides a means to identify enzyme inhibition, stability and equilibrium issues, or other potential bottlenecks, while increasing the substrate concentration in a stepwise manner. In case of biocatalytic reactions requiring cofactors and their regeneration, reducing system complexity and cofactor concentration are additional goals.
This has been demonstrated in the synthesis of the enantiomerically pure lactaldehydes by the reaction engineering of the enzymatic reduction of 1,1-dimethoxy-protected 2-propanone to (S)- and (R)-1,1-dimethoxy-2-propanols93 with the decrease of the NADP-concentration from 0.8 g per liter to 0.05 g per liter and the increase of the substrate concentration from 6 g per liter to 250 g per liter. For making available highly valuable metabolites in larger amounts biocatalytic reaction engineering towards achieving much lower substrate concentrations of the order of grams per liter can however also be of much interest. The biocatalytic 3-step conversion of α-linolenic acid to the plant metabolite cis-(+)-12-oxophytodienoic acid in one pot has been optimized to achieve >99% conversion, high diastereoselectivity and a reduction of side products by avoiding the decomposition of sensitive intermediates.94
Detailed analyses of the effects of reaction conditions on the kinetics and thermodynamics of the enzyme-catalyzed reactions and on stabilities of the reaction partners are essential for reaction optimization and reactor evaluation.95 Learning from dynamic analytical measurements of cellular metabolites by fast and sensitive mass spectrometry methods, well established in metabolomics, is important for improving process and product metrics by identifying where bottlenecks exist and where essential intermediates are lost.96
One of the key parameters of metabolite synthesis is related to the sustainability, not only of the reaction part of the process, as also the sustainability of the preceding and subsequent operations are relevant (see Fig. 7).
Therefore, in addition to the optimization of the reaction part, also the upstream and downstream parts need to be considered for the development of an intensified, scalable and viable biocatalytic process.97 The selected options in the upstream part, such as the choice of the whole-cell or cell-free system, can have a significant influence on the efficiency and effort going into the downstream operations for recovering and purifying the metabolite from the mixture after the reaction has been completed.98 Optimized product recovery and purification to the desired quality are essential for an overall process intensification and a high isolated yield of the final metabolite.99
Enzyme immobilization has been instrumental for increased enzyme stability and robustness and reduced reaction times, for example in the flow-based synthesis of melatonin from 5-methoxytryptamine catalyzed by immobilized acetyltransferase.100 The amount of enzyme and resin as well as the time required for screening and developing the immobilization of enzymes on affinity resins have been significantly reduced by automating the process development and optimization of enzyme immobilization.101
Scaling metabolite synthesis
The scaling of optimized batch processes, using chemical reactions, fermentation, whole-cell or cell-free biocatalytic processes, or combinations thereof, by their transfer from small-scale vessels via reactors of increasing volume has been key to the manufacturing of metabolites at the appropriate scale.102,103 Numerous processes for metabolite synthesis have been scaled up to a wide range of production scales, depending on the metabolite type, industrial sector and application. Small scale can be adequate for producing drug or research metabolites, but the time requirements and urgency of scaling may be the most important factors. Medium scale can be sufficient for routinely manufacturing batches of special metabolites with industrial applications. Industrial large scale has been well established in classical processes for the production of amino acids, organic acids, vitamins, nucleotides, steroids, vitamins, antibiotics and pharmaceutical metabolites. Their large scale production and global availability contributed tremendously to health, nutrition and the quality of life and represents also an important sector of the bioeconomy.104–108 While the classical scale-up of reactions in batch mode is still the prevailing industrial workflow for metabolite manufacturing processes, the strategic advantages of scaling by continuous flow processes have received much attention.109–112Outlook
The first phase in developing sustainable syntheses of required metabolites benefits from progress in various relevant areas, such as widening biocatalytic reaction platforms, increasing experimental reaction knowledge, new information technology tools and data-bases and methodological advances in route design and selection. Building short biocatalytic routes to complex metabolites and developing novel reactions with wider implications for sustainable chemistry are of much interest. Inspirations can originate from de novo design, enzyme engineering and from progress in elucidating enzyme functions in biosynthetic pathways to natural products113 and by discovering novel enzymes utilizing central metabolites as building blocks.114,115 Practical and simple in vitro prototyping approaches are attractive due to reduced complexity and are facilitated by advances in DNA synthesis and high-level expression of suitable enzymes. Fast and sensitive analytical methods with high information content are essential in the experimental phase of rapidly building, testing, learning and optimizing biocatalytic systems for metabolite synthesis. An integrated view of the reaction engineering, considering also the connections with upstream and downstream operations, can be valuable for developing a viable biocatalytic process, which can be intensified and scaled. The production scale at which metabolites are manufactured is depending on various technological factors as well as market demand. Increasing demand requires either the transfer to fixed batch reactors of larger volume, including additional process development time, or to increase the time of operation when using mobile continuous flow reactors.Conclusions
The synthesis of metabolites has always been a very important area for science, industry and society, and is today experiencing a renaissance. The replacement of extractive manufacturing methods from limited biological resources or of lengthy chemical routes with an unfavorable waste to product ratio, by sustainable methods is thereby highly important for scaling metabolite synthesis. Although numerous advances have enabled rapid development of metabolite synthesis from small to large scale, the number of metabolites without synthetic access and the great structural diversity of metabolites require major efforts. Thereby several metabolite-specific boundary conditions are important, such as resource efficiency, the manufacturing capability for the required amounts of metabolites while complying with sustainability goals, protection of endangered species and biodiversity issues. Their consideration is therefore essential in the design and selection of sustainable routes for metabolite synthesis. Benefits can be derived from the integration of biological and chemical domains in retrosynthetic analysis and from new approaches in computer-assisted synthesis planning. The growing interface between computational approaches, information technology and experimental advances in the characterization of enzymes involved in biosynthetic pathways, enzyme engineering and biocatalytic reaction platforms looks very promising for guiding the design and selection of sustainable routes for metabolite synthesis. Meaningful and sensitive analytical methods providing adequate information are essential for rapid prototyping and iterative DBTL cycles. Optimizing the entire workflow needs to include, in addition to the upstream part and reaction engineering, also the metabolite recovery and purification as essential part of sustainable metabolite manufacturing. With the advances in various tools and methodologies for route selection and reaction engineering there is a bright future in discovering and developing novel routes for sustainable metabolite synthesis.Author contributions
Roland Wohlgemuth: conceptualization; formal analysis; methodology; writing – original draft; writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
I would like to thank all my coworkers and cooperation partners as well as the COST organization for supporting COST Action CM1303 on Systems Biocatalysis.References
- S. L. Schreiber, Nat. Chem. Biol., 2005, 1, 64 CrossRef CAS PubMed.
- S. L. McKnight, Cold Spring Harbor Symp. Quant. Biol., 2011, 76, 403 CrossRef CAS PubMed.
- R. Wohlgemuth, Biotechnol. J., 2018, 13, 1700620 CrossRef PubMed.
- C. A. Hutchison III, R. Y. Chuang, V. N. Noskov, N. Assad-Garcia, T. J. Deerinck, M. H. Ellisman, J. Gill, K. Kannan, B. J. Karas, L. Ma, J. F. Pelletier, Z.-Q. Qi, R. A. Richter, E. A. Strychalski, L. Sun, Y. Suzuki, B. Tsvetanova, K. S. Wise, H. O. Smith, J. I. Glass, C. Merryman, D. G. Gibson and J. C. Venter, Science, 2016, 351(6280), aad6253 CrossRef PubMed.
- J. C. Venter, J. I. Glass, C. A. Hutchison III and S. Vashee, Cell, 2022, 185, 2708 CrossRef CAS PubMed.
- M. Kanehisa, Y. Sato and M. Kawashima, Protein Sci., 2022, 31(1), 47 CrossRef CAS PubMed.
- A. Chang, L. Jeske, S. Ulbrich, J. Hofmann, J. Koblitz, I. Schomburg, M. Neumann-Schaal, D. Jahn and D. Schomburg, Nucleic Acids Res., 2021, 49(D1), D498 CrossRef CAS PubMed.
- P. Bansal, A. Morgat, K. B. Axelsen, V. Muthukrishnan, E. Coudert, L. Aimo, N. Hyka-Nouspikel, E. Gasteiger, A. Kerhornou, T. Batista Neto, M. Pozzato, M.-C. Blatter, A. Ignatchenko, N. Redaschi and A. Bridge, Nucleic Acids Res., 2022, 50(D1), D693 CrossRef CAS PubMed.
- D. S. Wishart, Y. D. Feunang, A. Marcu, A. C. Guo, K. Liang, R. Vázquez-Fresno, T. Sajed, D. Johnson, C. Li, N. Karu and Z. Sayeeda, Nucleic Acids Res., 2018, 46(D1), D608 CrossRef CAS PubMed.
- D. S. Wishart, E. Oler, H. Peters, A. Guo, S. Girod, S. Han, S. Saha, V. W. Lui, M. LeVatte, V. Gautam, R. Kaddurah-Daouk and N. Karu, Nucleic Acids Res., 2023, 51, D611 CrossRef CAS PubMed.
- G. Brundtland, Report of the World Commission on Environment and Development: Our Common Future, United Nations General Assembly document A/42/427, 1987, https://sustainabledevelopment.un.org/content/documents/5987our-common-future.pdf.
- R. A. Sheldon, Chem. Ind., 1992, 23, 903 Search PubMed.
- A. Aguilar, T. Twardowski and R. Wohlgemuth, Biotechnol. J., 2019, 14(8), 1800638 CrossRef PubMed.
- R. Wohlgemuth, ChemSusChem, 2022, 15(9), e202200402 CrossRef CAS PubMed.
- Commission Recommendation (EU), 2022/2510 of 8.12.2022 establishing a European assessment framework for ‘safe and sustainable by design’ chemicals and materials, Official Journal of the European Union, 2022, L 325, 179 Search PubMed , https://eurlex.europa.eu/legalcontent/EN/TXT/?uri=CELEX:32022H2510.
- Á. Calvillo, T. Pellicer, M. Carnicer and A. Planas, Bioengineering, 2022, 9, 365 CrossRef PubMed.
- S. K. Schwechheimer, E. Y. Park, J. L. Revuelta, J. Becker and C. Wittmann, Appl. Microbiol. Biotechnol., 2016, 100, 2107 CrossRef CAS PubMed.
- F. Wöhler, Ann. Phys., 1828, 88, 253 CrossRef.
- K. C. Nicolaou, C. R. H. Hale, C. Nilewski and H. A. Ioannidou, Chem. Soc. Rev., 2012, 41, 5185 RSC.
- K. M. Hugentobler and E. M. Carreira, Acc. Chem. Res., 2021, 54, 890 CrossRef CAS PubMed.
- K. C. Nicolaou, D. Vourloumis, N. Winssinger and P. S. Baran, Angew. Chem., Int. Ed., 2000, 39, 44 CrossRef CAS PubMed.
- J. Zhang, L. G. Hansen, O. Gudich, K. Viehrig, L. M. M. Lassen, L. Schrübbers, K. B. Adhikari, P. Rubaszka, E. Carrasquer-Alvarez, L. Chen, V. D'Ambrosio, B. Lehka, A. K. Haidar, S. Nallapareddy, K. Giannakou, M. Laloux, D. Arsovska, M. A. K. Jørgensen, L. J. G. Chan, M. Kristensen, H. B. Christensen, S. Sudarsan, E. A. Stander, E. Baidoo, C. J. Petzold, T. Wulff, S. E. O'Connor, V. Courdavault, M. K. Jensen and J. D. Keasling, Nature, 2022, 609, 341 CrossRef CAS PubMed.
- E. J. Corey and X. M. Cheng, The logic of chemical synthesis, John Wiley & Sons, New York, 1989 Search PubMed.
- E. J. Corey and W. T. Wipke, Science, 1969, 166, 178 CrossRef CAS PubMed.
- B. Liu, B. Ramsundar, P. Kawthekar, J. Shi, J. Gomes, Q. L. Nguyen, S. Ho, J. Sloane, P. Wender and V. Pande, ACS Cent. Sci., 2017, 3, 1103 CrossRef CAS PubMed.
- C. W. Coley, L. Rogers, W. H. Green and K. F. Jensen, ACS Cent. Sci., 2017, 3, 1237 CrossRef CAS PubMed.
- B. Mikulak-Klucznik, P. Gołębiowska, A. A. Bayly, O. Popik, T. Klucznik, S. Szymkuć, E. P. Gajewska, P. Dittwald, O. Staszewska-Krajewska, W. Beker, T. Badowski, K. A. Scheidt, K. Molga, J. Mlynarski, M. Mrksich and B. A. Grzybowski, Nature, 2020, 588, 83 CrossRef CAS PubMed.
- P. Schwaller, R. Petraglia, V. Zullo, V. H. Nair, R. A. Haeuselmann, R. Pisoni, C. Bekas, A. Iuliano and T. Laino, Chem. Sci., 2020, 11, 3316 RSC.
- Retrosynthesis at https://scifinder-n.cas.org/.
- M. H. S. Segler, M. Preuss and M. P. Waller, Nature, 2018, 555, 604 CrossRef CAS PubMed.
- R. Wohlgemuth, Biotechnol. J., 2009, 4, 1253 CrossRef CAS PubMed.
- N. J. Turner and E. O'Reilly, Nat. Chem. Biol., 2013, 9, 285 CrossRef CAS PubMed.
- A. P. Green and N. J. Turner, Perspect. Sci., 2016, 9, 42 CrossRef.
- R. O. de Souza, L. S. Miranda and U. T. Bornscheuer, Chem. – Eur. J., 2017, 23(50), 12040 CrossRef CAS PubMed.
- M. Hönig, P. Sondermann, N. J. Turner and E. M. Carreira, Angew. Chem., Int. Ed., 2017, 56(31), 8942 CrossRef PubMed.
- W. Finnigan, L. J. Hepworth, S. L. Flitsch and N. J. Turner, Nat. Catal., 2021, 4(2), 98 CrossRef CAS PubMed.
- D. Probst, M. Manica, Y. G. Nana Teukam, A. Castrogiovanni, F. Paratore and T. Laino, Nat. Commun., 2022, 13, 964 CrossRef CAS PubMed.
- H. MohammadiPeyhani, J. Hafner, A. Sveshnikova, V. Viterbo and V. Hatzimanikatis, Nat. Commun., 2022, 13, 1560 CrossRef CAS PubMed.
- B. Delépine, T. Duigou, P. Carbonell and J. L. Faulon, Metab. Eng., 2018, 45, 158 CrossRef PubMed.
- T. Yu, A. G. Boob, M. J. Volk, X. Liu, H. Cui and H. Zhao, Nat. Catal., 2023, 1–15, DOI:10.1038/s41929-022-00909-w.
- M. C. de la Torre and M. A. Sierra, Angew. Chem., Int. Ed., 2004, 43(2), 160 CrossRef CAS PubMed.
- M. Razzak and J. K. De Brabander, Nat. Chem. Biol., 2011, 7, 865 CrossRef CAS PubMed.
- R. Bao, H. Zhang and Y. Tang, Acc. Chem. Res., 2021, 54, 3720 CrossRef CAS PubMed.
- C. I. Lin, R. M. McCarty and H. W. Liu, Angew. Chem., Int. Ed., 2017, 56(13), 3446 CrossRef CAS PubMed.
- A. F. Kiefer, Y. C. Liu, R. Gummerer, C. Jäger and J. Deska, Angew. Chem., Int. Ed., 2023, e202301178 CAS.
- B. E. Hetzler, D. Trauner and A. L. Lawrence, Nat. Rev. Chem., 2022, 6, 170 CrossRef PubMed.
- I. Levin, M. Liu, C. A. Voigt and C. W. Coley, Nat. Commun., 2022, 13, 7747 CrossRef CAS PubMed.
- K. Sankaranarayanan, E. Heid, C. W. Coley, D. Verma, W. H. Green and K. F. Jensen, Chem. Sci., 2022, 13, 6039 RSC.
- R. Wohlgemuth, T. Twardowski and A. Aguilar, New Biotechnol., 2021, 61, 22–28 CrossRef CAS PubMed.
- R. Wohlgemuth, Chem. Biochem. Eng. Q., 2017, 31, 131–138 CrossRef CAS.
- G. B. Kim, S. Y. Choi, I. J. Cho, D. H. Ahn and S. Y. Lee, Trends Biotechnol., 2023, 41(3), 425 CrossRef CAS PubMed.
- C. A. Voigt, Nat. Commun., 2020, 11, 6379 CrossRef CAS PubMed.
- S. J. Moore, J. T. MacDonald and P. S. Freemont, Biochem. Soc. Trans., 2017, 45, 785 CrossRef CAS PubMed.
- A. Casini, F. Y. Chang, R. Eluere, A. M. King, E. M. Young, Q. M. Dudley, A. Karim, K. Pratt, C. Bristol, A. Forget, A. Ghodasara, R. Warden-Rothman, R. Gan, A. Cristofaro, A. E. Borujeni, M. H. Ryu, J. Li, Y. C. Kwon, H. Wang, E. Tatsis, C. Rodriguez-Lopez, S. O'Connor, M. H. Medema, M. A. Fischbach, M. C. Jewett, C. Voigt and D. B. Gordon, J. Am. Chem. Soc., 2018, 140, 4302–4316 CrossRef CAS PubMed.
- P. Carbonell, A. J. Jervis, C. J. Robinson, C. Yan, M. Dunstan, N. Swainston, M. Vinaixa, K. A. Hollywood, A. Currin, N. J. W. Rattray, S. Taylor, R. Spiess, R. Sung, A. R. Williams, D. Fellows, N. J. Stanford, P. Mulherin, R. Le Feuvre, P. Barran, R. Goodacre, N. J. Turner, C. Goble, G. G. Chen, D. B. Kell, J. Micklefield, R. Breitling, E. Takano, J. L. Faulon and N. S. Scrutton, Commun. Biol., 2018, 1, 66 CrossRef PubMed.
- J. Nielsen and J. D. Keasling, Cell, 2016, 164, 1185 CrossRef CAS PubMed.
- A. H. Singh, B. B. Kaufmann-Malaga, J. A. Lerman, D. P. Dougherty, Y. Zhang, A. L. Kilbo, E. H. Wilson, C. Y. Ng, O. Erbilgin, K. A. Curran, C. D. Reeves, J. E. Hung, S. Mantovani, Z. A. King, M. J. Ayson, J. R. Denery, C. W. Lu, P. Norton, C. Tran, D. M. Platt, J. R. Cherry, S. S. Chandran and A. L. Meadows, bioRxiv, 2023, preprint, DOI:10.1101/2023.01.03.521657.
- X. Liao, H. Ma and Y. J. Tang, Curr. Opin. Biotechnol., 2022, 75, 102712 CrossRef CAS PubMed.
- COST Action CM1303 - Systems Biocatalysis, https://www.cost.eu/actions/CM1303/.
- Systems biocatalysis for bioprocess design, ed. R. Wohlgemuth, J. A. Littlechild and B. G. Kim, Frontiers Media SA, 2022 Search PubMed.
- L. Caputi, J. Franke, S. C. Farrow, K. Chung, R. M. Payne, T. D. Nguyen, T. T. T. Dang, I. Soares Teto Carqueijeiro, K. Koudounas, T. Dugé de Bernonville, B. Ameyaw, D. M. Jones, I. J. Curcino Vieira, V. Courdavault and S. E. O'Connor, Science, 2018, 360(6394), 1235 CrossRef CAS PubMed.
- D. Yang, W. D. Jang and S. Y. Lee, J. Am. Chem. Soc., 2021, 143, 5364 CrossRef CAS PubMed.
- J. R. Swartz, Metab. Eng., 2018, 50, 156 CrossRef CAS PubMed.
- J. W. Bogart, M. D. Cabezas, B. Vögeli, D. A. Wong, A. S. Karim and M. C. Jewett, ChemBioChem, 2021, 22(1), 84 CrossRef CAS PubMed.
- X. Ji, W. Q. Liu and J. Li, Curr. Opin. Microbiol., 2022, 67, 102142 CrossRef CAS PubMed.
- A. Hoose, R. Vellacott, M. Storch, P. S. Freemont and M. G. Ryadnov, Nat. Rev. Chem., 2023, 7, 144 CrossRef CAS PubMed.
- Cell-free Protein Synthesis: Methods and Protocols, ed. A. S. Spirin and J. R. Swartz, Wiley-VCH, Weinheim, Germany, 2008 Search PubMed.
- L. Jiang, J. Zhao, J. Lian and Z. Xu, Synth. Syst. Biotechnol., 2018, 3(2), 90 CrossRef PubMed.
- A. D. Silverman, A. S. Karim and M. C. Jewett, Nat. Rev. Genet., 2020, 21(3), 151 CrossRef CAS PubMed.
- A. S. Karim and M. C. Jewett, Metab. Eng., 2016, 36, 116 CrossRef CAS PubMed.
- Q. M. Dudley, A. S. Karim, C. J. Nash and M. C. Jewett, Metab. Eng., 2020, 61, 251 CrossRef CAS PubMed.
- B. J. Rasor, B. Vögeli, M. C. Jewett and A. S. Karim, Cell-Free Gene Expression: Methods and Protocols, 2022, p. 199 Search PubMed.
- K. Haslinger, T. Hackl and K. L. J. Prather, Cell Chem. Biol., 2021, 28(6), 876 CrossRef CAS PubMed.
- K. Matsubara, R. Köhling, B. Schönenberger, T. Kouril, D. Esser, C. Bräsen, B. Siebers and R. Wohlgemuth, J. Biotechnol., 2014, 191, 69 CrossRef CAS PubMed.
- B. Schoenenberger, A. Wszolek, T. Milesi, H. Brundiek, M. Obkircher and R. Wohlgemuth, ChemCatChem, 2017, 9(1), 121 CrossRef CAS.
- B. Schoenenberger, A. Wszolek, R. Meier, H. Brundiek, M. Obkircher and R. Wohlgemuth, RSC Adv., 2017, 7, 48952 RSC.
- N. Hardt, B. M. Kinfu, J. Chow, B. Schoenenberger, W. R. Streit, M. Obkircher and R. Wohlgemuth, ChemBioChem, 2017, 18(15), 1518 CrossRef CAS PubMed.
- B. Schoenenberger, A. Wszolek, R. Meier, H. Brundiek, M. Obkircher and R. Wohlgemuth, Biotechnol. J., 2018, 13(8), 1700529 CrossRef PubMed.
- N. Hardt, S. Kind, B. Schoenenberger, T. Eggert, M. Obkircher and R. Wohlgemuth, Biocatal. Biotransform., 2020, 38(1), 35 CrossRef CAS.
- B. Schoenenberger, S. Kind, R. Meier, T. Eggert, M. Obkircher and R. Wohlgemuth, Biocatal. Biotransform., 2020, 38(1), 53 CrossRef CAS.
- A. O. Chatzivasileiou, V. Ward, S. McBride Edgar and G. Stephanopoulos, Proc. Natl. Acad. Sci. U. S. A., 2019, 116(2), 506 CrossRef CAS PubMed.
- C. J. Paddon and J. D. Keasling, Nat. Rev. Microbiol., 2014, 12(5), 355 CrossRef CAS PubMed.
- J. Zhang, L. G. Hansen, O. Gudich, K. Viehrig, L. M. Lassen, L. Schrübbers, K. B. Adhikari, P. Rubaszka, E. Carrasquer-Alvarez, L. Chen, V. D'Ambrosio, B. Lehka, A. K. Haidar, S. Nallapareddy, K. Giannakou, M. Laloux, D. Arsovska, M. A. K. Jørgensen, L. J. G. Chan, M. Kristensen, H. B. Christensen, S. Sudarsan, E. A. Stander, E. Baidoo, C. J. Petzold, T. Wulff, S. E. O'Connor, V. Courdavault, M. K. Jensen and J. D. Keasling, Nature, 2022, 609(7926), 341 CrossRef CAS PubMed.
- C. A. Roessner, J. B. Spencer, N. J. Stolowich, J. Wang, G. P. Nayar, P. J. Santander, C. Pichon, C. Min, M. T. Holderman and A. I. Scott, Chem. Biol., 1994, 1(2), 119 CrossRef CAS PubMed.
- L. Shen, M. Kohlhaas, J. Enoki, R. Meier, B. Schönenberger, R. Wohlgemuth, R. Kourist, F. Niemeyer, D. van Niekerk, C. Bräsen, J. Niemeyer, J. Snoep and B. Siebers, Nat. Commun., 2020, 11(1), 1098 CrossRef CAS PubMed.
- A. S. Karim, Q. M. Dudley, A. Juminaga, Y. Yuan, S. A. Crowe, J. T. Heggestad, S. Garg, T. Abdalla, W. S. Grubbe, B. J. Rasor, D. N. Coar, M. Torculas, M. Krein, F. Liew, A. Quattlebaum, R. O. Jensen, J. A. Stuart, S. D. Simpson, M. Köpke and M. C. Jewett, Nat. Chem. Biol., 2020, 16, 912 CrossRef CAS PubMed.
- J. W. Bogart, M. D. Cabezas, B. Vögeli, D. A. Wong, A. S. Karim and M. C. Jewett, ChemBioChem, 2021, 22(1), 84 CrossRef CAS PubMed.
- M. Teshima, V. P. Willers and V. Sieber, Curr. Opin. Biotechnol., 2023, 79, 102868 CrossRef CAS PubMed.
- J. M. Woodley, React. Chem. Eng., 2020, 5, 632 RSC.
- N. J. Claassens, S. Burgener, B. Vögeli, T. J. Erb and A. Bar-Even, Curr. Opin. Biotechnol., 2019, 60, 221 CrossRef CAS PubMed.
- C. Katsimpouras and G. Stephanopoulos, Curr. Opin. Biotechnol., 2021, 69, 91 CrossRef CAS PubMed.
- R. A. Sheldon, Green Chem., 2023, 25, 1704 RSC.
- M. A. K. Vogel, H. Burger, N. Schläger, R. Meier, B. Schönen-berger, T. Bisschops and R. Wohlgemuth, React. Chem. Eng., 2016, 1(2), 156 RSC.
- J. Löwe, K. J. Dietz and H. Gröger, Adv. Sci., 2020, 7, 1902973 CrossRef PubMed.
- G. S. Molla, B. M. Kinfu, J. Chow, W. Streit, R. Wohlgemuth and A. Liese, Biotechnol. J., 2017, 12(3), 1600625 CrossRef PubMed.
- C. J. Vavricka, T. Hasunuma and A. Kondo, Trends Biotechnol., 2020, 38(1), 68 CrossRef CAS PubMed.
- K. V. K. Boodhoo, M. C. Flickinger, J. M. Woodley and E. A. C. Emanuelsson, Chem. Eng. Process., 2022, 172, 108793 CrossRef CAS.
- R. Wohlgemuth, Biotechnol. J., 2009, 4(9), 1253 CrossRef CAS PubMed.
- R. Wohlgemuth, in Comprehensive Biotechnology, Elsevier, Amsterdam, The Netherlands, 2011, pp. 591–601 Search PubMed.
- M. L. Contente, S. Farris, L. Tamborini, F. Molinari and F. Paradisi, Green Chem., 2019, 21, 3263 RSC.
- J. H. Forstater and S. T. Grosser, React. Chem. Eng., 2022, 7, 866 RSC.
- A. L. Demain, Biotechnol. Adv., 2000, 18(6), 499–514 CrossRef CAS PubMed.
- Industrial Biotrans-formations, ed. A. Liese, K. Seelbach and C. Wandrey, Wiley-VCH, Weinheim, Germany, 2nd edn, completely revised and extended edition, 2006 Search PubMed.
- Industrial Biotechnology: Sustainable Growth and Economic Success, ed. W. Soetaert and E. J. Vandamme, Wiley-VCH, Weinheim, Germany, 2010 Search PubMed.
- Industrial Biotechnology: Products and Processes, ed. C. Wittmann, J. C. Liao, S. Y. Lee, J. Nielsen and G. Stephanopoulos, Wley-VCH, Weinheim, Germany, 2017 Search PubMed.
- S. Sanchez, R. Rodríguez-Sanoja, A. Ramos and A. L. Demain, J. Antibiot., 2018, 71(1), 26 CrossRef CAS PubMed.
- B. Hoff, J. Plassmeier, M. Blankschien, A. C. Letzel, L. Kourtz, H. Schröder, W. Koch and O. Zelder, Angew. Chem., Int. Ed., 2021, 60(5), 2258 CrossRef CAS PubMed.
- M. Eggersdorfer, D. Laudert, U. Letinois, T. McClymont, J. Medlock, T. Netscher and W. Bonrath, Angew. Chem., Int. Ed., 2012, 51(52), 12960 CrossRef CAS PubMed.
- R. Wohlgemuth, I. Plazl, P. Žnidaršič-Plazl, K. V. Gernaey and J. M. Woodley, Trends Biotechnol., 2015, 33(5), 302 CrossRef CAS PubMed.
- P. Gruber, M. P. Marques, B. O'Sullivan, F. Baganz, R. Wohlgemuth and N. Szita, Biotechnol. J., 2017, 12(7), 1700030 CrossRef PubMed.
- L. Tamborini, P. Fernandes, F. Paradisi and F. Molinari, Trends Biotechnol., 2018, 36(1), 73 CrossRef CAS PubMed.
- P. De Santis, L. E. Meyer and S. Kara, React. Chem. Eng., 2020, 5(12), 2155 RSC.
- C. T. Walsh, Nat. Prod. Rep., 2023, 40, 326 RSC.
- L. Barra, T. Awakawa and I. Abe, JACS Au, 2022, 2(9), 1950 CrossRef CAS PubMed.
- Y. H. Lee, D. Ren, B. Jeon and H. w. Liu, Nat. Prod. Rep., 2023 10.1039/d2np00086e.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3re00222e |
| This journal is © The Royal Society of Chemistry 2023 |